Abstract
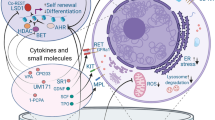
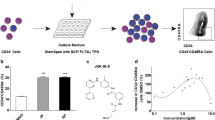
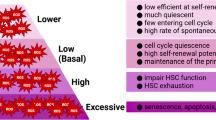
Hematopoietic stem cell (HSCs) transplantation is the primary therapeutic modality used to treat hematopoietic disorders. It centers on the capability of a small quantity of HSCs to repopulate whole blood lineages. Along with limited availability of suitable donors, the need for sufficient number of donor HSCs is still challenging in clinical relevance. This has been addressed by ex vivo HSC expansion albeit with partial success, and thus development of an alternative strategy that could improve HSC expansion is required. To that end, we aimed to build HematoMiR, an oligo-based technology that broadly targets HSC quiescence factors. Here, we show that HematoMiRs and their combinations targeting over 50 factors involved in HSC quiescence could induce robust ex vivo murine and human HSC expansion. In particular, HematoMiR-5 treatment enhanced cell cycle through down-regulation of negative cell cycle regulators in HSCs. HematoMiR-5 treated HSPCs had reduced DNA damage during the course of ex vivo expansion. Moreover, HematoMiR-5 treatment led to sustained HSC self-renewal ability and a low apoptosis rate. In addition, HematoMiR-5 expanded HSCs demonstrated successful engraftment and repopulation capacity in the recipient animals. Furthermore, combinatorial treatments of HematoMiR-2 and 5 allowed vigorous ex vivo HSC expansion. These findings demonstrate that novel and synthetic HematoMiR technology is feasible for HSC ex vivo expansion through the sequence-dependent modulation of numerous HSC quiescence modulators.
Graphical abstract














Similar content being viewed by others
Data availability
The data that supports the findings of this study are available in the supplementary material of this article.
References
Yucel D, Kocabas F (2017) Developments in hematopoietic stem cell expansion and gene editing technologies. Adv Exp Med Biol 1079:103–125. https://doi.org/10.1007/5584_2017_114
Iscove NN, Nawa K (1997) Hematopoietic stem cells expand during serial transplantation in vivo without apparent exhaustion. Curr Biol 7:805–808. https://doi.org/10.1016/S0960-9822(06)00341-1
Matatall KA et al (2016) Chronic infection depletes hematopoietic stem cells through stress-induced terminal differentiation. Cell Rep 17:2584–2595. https://doi.org/10.1016/j.celrep.2016.11.031
Pawliuk R, Eaves C, Humphries RK (1996) Evidence of both ontogeny and transplant dose-regulated expansion of hematopoietic stem cells in vivo. Blood 88:2852–2858. https://doi.org/10.1182/blood.V88.8.2852.bloodjournal8882852
Jansen J, Hanks S, Thompson JM, Dugan MJ, Akard LP (2005) Transplantation of hematopoietic stem cells from the peripheral blood. J Cell Mol Med 9:37–50. https://doi.org/10.1111/j.1582-4934.2005.tb00335.x
Domen J, Weissman IL (1999) Self-renewal, differentiation or death: regulation and manipulation of hematopoietic stem cell fate. Mol Med Today 5:201–208. https://doi.org/10.1016/S1357-4310(99)01464-1
Uchida N et al (1998) High doses of purified stem cells cause early hematopoietic recovery in syngeneic and allogeneic hosts. J Clin Investig 101:961–966
Stevens CE, Scaradavou A, Carrier C, Carpenter C, Rubinstein P (2005) An empirical analysis of the probability of finding a well matched cord blood unit: implications for a national cord blood inventory. Blood 106:2047–2047. https://doi.org/10.1182/blood.V106.11.2047.2047
Remberger M et al (2015) Effect of total nucleated and CD34(+) cell dose on outcome after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transpl 21:889–893. https://doi.org/10.1016/j.bbmt.2015.01.025
Broxmeyer HE (2016) Enhancing the efficacy of engraftment of cord blood for hematopoietic cell transplantation. Transfus Apher Sci 54:364–372. https://doi.org/10.1016/j.transci.2016.05.013
Kim YJ, Broxmeyer HE (2011) Immune regulatory cells in umbilical cord blood and their potential roles in transplantation tolerance. Crit Rev Oncol Hematol 79:112–126. https://doi.org/10.1016/j.critrevonc.2010.07.009
Zheng J et al (2011) Ex vivo expanded hematopoietic stem cells overcome the MHC barrier in allogeneic transplantation. Cell Stem Cell 9:119–130. https://doi.org/10.1016/j.stem.2011.06.003
Von Drygalski A, Ren L, Adamson JW (2004) Murine bone marrow cells cultured ex vivo in the presence of multiple cytokine combinations lose radioprotective and long-term engraftment potential. Stem Cells Dev 13:101–111
Audet J, Miller CL, Eaves CJ, Piret JM (2002) Common and distinct features of cytokine effects on hematopoietic stem and progenitor cells revealed by dose-response surface analysis. Biotechnol Bioeng 80:393–404. https://doi.org/10.1002/bit.10399
Derakhshani M et al (2019) Strategies for elevating hematopoietic stem cells expansion and engraftment capacity. Life Sci 232:116598. https://doi.org/10.1016/j.lfs.2019.116598
Kumar S, Geiger H (2017) HSC niche biology and HSC expansion ex vivo. Trends Mol Med 23:799–819. https://doi.org/10.1016/j.molmed.2017.07.003
Walasek MA, van Os R, de Haan G (2012) Hematopoietic stem cell expansion: challenges and opportunities. Ann N Y Acad Sci 1266:138–150. https://doi.org/10.1111/j.1749-6632.2012.06549.x
Yucel D, Kocabas F (2017) Developments in hematopoietic stem cell expansion and gene editing technologies. Adv Exp Med Biol. https://doi.org/10.1007/5584_2017_114
Takubo K et al (2013) Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12:49–61. https://doi.org/10.1016/j.stem.2012.10.011
Perry JM et al (2011) Cooperation between both Wnt/β-catenin and PTEN/PI3K/Akt signaling promotes primitive hematopoietic stem cell self-renewal and expansion. Genes Dev 25:1928–1942. https://doi.org/10.1101/gad.17421911
Uslu M, Albayrak E, Kocabaş F (2020) Temporal modulation of calcium sensing in hematopoietic stem cells is crucial for proper stem cell expansion and engraftment. J Cell Physiol. https://doi.org/10.1002/jcp.29777
Croci S et al (2019) (64)Cu and fluorescein labeled anti-miRNA peptide nucleic acids for the detection of miRNA expression in living cells. Sci Rep 9:3376–3376. https://doi.org/10.1038/s41598-018-35800-x
Diener Y et al (2015) RNA-based, transient modulation of gene expression in human haematopoietic stem and progenitor cells. Sci Rep 5:17184. https://doi.org/10.1038/srep17184
Turan RD et al (2020) Development of small molecule meis inhibitors that modulate HSC activity. Sci Rep. https://doi.org/10.1038/s41598-020-64888-3
Uslu M, Albayrak E, Kocabaş F (2020) Temporal modulation of calcium sensing in hematopoietic stem cells is crucial for proper stem cell expansion and engraftment. J Cell Physiol 235:9644–9666. https://doi.org/10.1002/jcp.29777
Aksoz M et al (2019) c-Myc inhibitor 10074–G5 induces murine and human hematopoietic stem and progenitor cell expansion and HDR modulator Rad51 expression. Curr Cancer Drug Targets 19:479–494. https://doi.org/10.2174/1568009618666180905100608
Kocabas F, Zheng J, Zhang C, Sadek HA (2014) Metabolic characterization of hematopoietic. Stem Cells 1185:155–164. https://doi.org/10.1007/978-1-4939-1133-2_10
Kocabas F et al (2012) Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood 120:4963–4972. https://doi.org/10.1182/blood-2012-05-432260
Simsek T et al (2010) The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7:380–390. https://doi.org/10.1016/j.stem.2010.07.011
Kocabas F et al (2015) Hypoxic metabolism in human hematopoietic stem cells. Cell Biosci. https://doi.org/10.1186/s13578-015-0020-3
Zheng J et al (2014) Profilin 1 is essential for retention and metabolism of mouse hematopoietic stem cells in bone marrow. Blood 123:992–1001. https://doi.org/10.1182/blood-2013-04-498469
Agarwal V, Bell GW, Nam J-W, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:e05005. https://doi.org/10.7554/eLife.05005
Kozomara A, Birgaoanu M, Griffiths-Jones S (2018) miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162. https://doi.org/10.1093/nar/gky1141
Hsu S-D et al (2010) miRTarBase: a database curates experimentally validated microRNA–target interactions. Nucleic Acids Res 39:D163–D169. https://doi.org/10.1093/nar/gkq1107
Vlachos IS et al (2012) DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res 40:W498–W504. https://doi.org/10.1093/nar/gks494
Sticht C, De La Torre C, Parveen A, Gretz N (2018) miRWalk: an online resource for prediction of microRNA binding sites. PLoS ONE 13:e0206239. https://doi.org/10.1371/journal.pone.0206239
Xiao F et al (2008) miRecords: an integrated resource for microRNA–target interactions. Nucleic Acids Res 37:D105–D110. https://doi.org/10.1093/nar/gkn851
Mahmoud AI et al (2013) Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature 497:249–253. https://doi.org/10.1038/nature12054
van Velthoven CTJ, Rando TA (2019) Stem cell quiescence: dynamism, restraint, and cellular idling. Cell Stem Cell 24:213–225. https://doi.org/10.1016/j.stem.2019.01.001
Orford KW, Scadden DT (2008) Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet 9:115. https://doi.org/10.1038/nrg2269
Wilson A, Laurenti E, Trumpp A (2009) Balancing dormant and self-renewing hematopoietic stem cells. Curr Opin Genet Dev 19:461–468. https://doi.org/10.1016/j.gde.2009.08.005
Yamada T, Park CS, Lacorazza HD (2013) Genetic control of quiescence in hematopoietic stem cells. Cell Cycle 12:2376–2383. https://doi.org/10.4161/cc.25416
Milsom MD (2019) Ex vivo expansion of functional hematopoietic stem cells, facilitating transplantation in the absence of conditioning. Hemasphere 3:e306–e306. https://doi.org/10.1097/HS9.0000000000000306
Dykxhoorn DM, Novina CD, Sharp PA (2003) Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol 4:457–467. https://doi.org/10.1038/nrm1129
Lam JKW, Chow MYT, Zhang Y, Leung SWS (2015) siRNA versus miRNA as therapeutics for gene silencing. Mol Therapy Nucleic Acids 4:e252. https://doi.org/10.1038/mtna.2015.23
Agrawal N et al (2003) RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev 67:657–685. https://doi.org/10.1128/MMBR.67.4.657-685.2003
Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524. https://doi.org/10.1038/nrm3838
Lim LP et al (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773. https://doi.org/10.1038/nature03315
Bader AG, Brown D, Stoudemire J, Lammers P (2011) Developing therapeutic microRNAs for cancer. Gene Ther 18:1121–1126. https://doi.org/10.1038/gt.2011.79
van Rooij E, Kauppinen S (2014) Development of microRNA therapeutics is coming of age. EMBO Mol Med 6:851–864. https://doi.org/10.15252/emmm.201100899
Yilmaz ÖH et al (2006) Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441:475–482. https://doi.org/10.1038/nature04703
Zhang J et al (2006) PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441:518–522. https://doi.org/10.1038/nature04747
Menon V, Ghaffari S (2018) Transcription factors FOXO in the regulation of homeostatic hematopoiesis. Curr Opin Hematol 25:290–298. https://doi.org/10.1097/MOH.0000000000000441
Miyamoto K et al (2007) Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1:101–112. https://doi.org/10.1016/j.stem.2007.02.001
Tothova Z et al (2007) FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128:325–339. https://doi.org/10.1016/j.cell.2007.01.003
Yamazaki S et al (2006) Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. EMBO J 25:3515–3523. https://doi.org/10.1038/sj.emboj.7601236
Dijkers PF et al (2000) Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol Cell Biol 20:9138–9148. https://doi.org/10.1128/mcb.20.24.9138-9148.2000
Medema RH, Kops GJPL, Bos JL, Burgering BMT (2000) AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782–787. https://doi.org/10.1038/35008115
Liu Y et al (2009) p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4:37–48. https://doi.org/10.1016/j.stem.2008.11.006
Naka K, Muraguchi T, Hoshii T, Hirao A (2008) Regulation of reactive oxygen species and genomic stability in hematopoietic stem cells. Antioxid Redox Signal 10:1883–1894. https://doi.org/10.1089/ars.2008.2114
Richardson C, Stark JM, Ommundsen M, Jasin M (2004) Rad51 overexpression promotes alternative double-strand break repair pathways and genome instability. Oncogene 23:546–553. https://doi.org/10.1038/sj.onc.1207098
Wright WD, Shah SS, Heyer W-D (2018) Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem 293:10524–10535. https://doi.org/10.1074/jbc.TM118.000372
Post S et al (2001) Phosphorylation of serines 635 and 645 of human Rad17 is cell cycle regulated and is required for G1/S checkpoint activation in response to DNA damage. Proc Natl Acad Sci 98:13102. https://doi.org/10.1073/pnas.231364598
Post SM, Tomkinson AE, Lee EYHP (2003) The human checkpoint Rad protein Rad17 is chromatin-associated throughout the cell cycle, localizes to DNA replication sites, and interacts with DNA polymerase epsilon. Nucleic Acids Res 31:5568–5575. https://doi.org/10.1093/nar/gkg765
Filippi M-D, Ghaffari S (2019) Mitochondria in the maintenance of hematopoietic stem cells: new perspectives and opportunities. Blood 133:1943–1952. https://doi.org/10.1182/blood-2018-10-808873
Rossi DJ et al (2007) Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447:725–729. https://doi.org/10.1038/nature05862
Zou G-M, Yoder MC (2005) Application of RNA interference to study stem cell function: current status and future perspectives. Biol Cell 97:211–219. https://doi.org/10.1042/bc20040084
Besharat ZM et al (2018) Low expression of miR-466f-3p sustains epithelial to mesenchymal transition in sonic hedgehog medulloblastoma stem cells through Vegfa-Nrp2 signaling pathway. Front Pharmacol. https://doi.org/10.3389/fphar.2018.01281
Chen YQ et al (2012) Abated microRNA-195 expression protected mesangial cells from apoptosis in early diabetic renal injury in mice. J Nephrol 25:566–576. https://doi.org/10.5301/jn.5000034
Hunsberger JG, Fessler EB, Wang Z, Elkahloun AG, Chuang DM (2012) Post-insult valproic acid-regulated microRNAs: potential targets for cerebral ischemia. Am J Transl Res 4:316–332
Ding Y et al (2012) Expression profile of miRNAs in APP swe/PSΔE9 transgenic mice. Nan Fang Yi Ke Da Xue Xue Bao 32:1280–1283
An F et al (2012) miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis 17:702–716. https://doi.org/10.1007/s10495-012-0704-7
Gong Y et al (2014) MicroRNA expression signature for Satb2-induced osteogenic differentiation in bone marrow stromal cells. Mol Cell Biochem 387:227–239. https://doi.org/10.1007/s11010-013-1888-z
Minwen Xu, Wang Liefeng Su, Dong-Ming. (2017) A fine-tune role of Mir-125a-5p on Foxn1 during age-associated changes in the thymus. Aging Dis 8:277–286. https://doi.org/10.14336/ad.2016.1109
Zhang Q et al (2018) Maternal chromium restriction induces insulin resistance in adult mice offspring through miRNA. Int J Mol Med 41:1547–1559. https://doi.org/10.3892/ijmm.2017.3328
Stiles JM, Kurisetty V, Mitchell DC, Bryan BA (2013) Rho kinase proteins regulate global miRNA expression in endothelial cells. Cancer Genom Proteom 10:251–263
Chen J et al (2017) Interaction of RNA-binding protein HuR and miR-466i regulates GM-CSF expression. Sci Rep 7:17233. https://doi.org/10.1038/s41598-017-17371-5
Mukherjee B et al (2015) Antimony-resistant Leishmania donovani exploits miR-466i to deactivate host MyD88 for regulating IL-10/IL-12 levels during early hours of infection. J Immunol 195:2731–2742. https://doi.org/10.4049/jimmunol.1402585
Li Z-Y, Na H-M, Peng G, Pu J, Liu P (2011) Alteration of microRNA expression correlates to fatty acid-mediated insulin resistance in mouse myoblasts. Mol BioSyst 7:871–877. https://doi.org/10.1039/C0MB00230E
Sunwoo J-S et al (2018) Maternal immune activation alters brain microRNA expression in mouse offspring. Ann Clin Transl Neurol 5:1264–1276. https://doi.org/10.1002/acn3.652
Bai T et al (2019) Expansion of primitive human hematopoietic stem cells by culture in a zwitterionic hydrogel. Nat Med 25:1566–1575. https://doi.org/10.1038/s41591-019-0601-5
Wilkinson AC et al (2019) Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature 571:117–121. https://doi.org/10.1038/s41586-019-1244-x
Acknowledgements
We like to thank Dr. Emrah Nikerel for initial automations. We would like to thank to Dr. Esra Albayrak and Dr. Rukset Attar (UCB sample collection), Dr. Esra Albayrak (animal handling in retro-orbital injections), Dr. Neslihan Meric and Dr. Zafer Gulbas (BM sample collection), Dr. Safa Aydin (technical assistance of using electroporater).
Funding
We thank the support from The International Centre for Genetic Engineering and Biotechnology—ICGEB 2015 Early Career Return Grant [grant number CRP/TUR15-02_EC]. MU has been supported by TÜBİTAK 215Z071 and 118S540.
Author information
Authors and Affiliations
Contributions
MU performed the experiments, collected the data, prepared the figures, and wrote the draft of the article. FK designed the experiments, contributed to data analysis, figure preparations and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest concerning this work.
Ethical approvals
All animal studies were approved with ethical permission from Institutional Animal Care and Use Committee of Yeditepe University (YUDHEK) with the decision number 651. All human studies were performed with consent to participate under the approval given by the Institutional Clinical Studies Ethical Committee of Yeditepe University with the permission decision numbers: 1067 and 944.
Consent for publication
All authors have contributed to this manuscript, reviewed and approved the current form of the manuscript to be submitted.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Uslu, M., Kocabaş, F. Development of a novel and synthetic HematoMiR technology that broadly modulates quiescence of stem cells and enhances HSC expansion. Cell. Mol. Life Sci. 79, 68 (2022). https://doi.org/10.1007/s00018-021-04031-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-021-04031-1