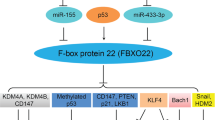
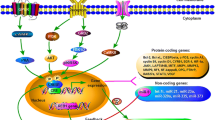
Abstract
HMG box protein 1 (HBP1) is a transcription factor and a potent cell cycle inhibitor in normal and cancer cells. HBP1 activates or represses the expression of different cell cycle genes (such as CDKN2A, CDKN1A, and CCND1) through direct DNA binding, cofactor recruitment, chromatin remodeling, or neutralization of other transcription factors. Among these are LEF1, TCF4, and MYC in the WNT/beta-catenin pathway. HBP1 also contributes to oncogenic RAS-induced senescence and terminal cell differentiation. Collectively, these activities suggest a tumor suppressor function. However, HBP1 is not listed among frequently mutated cancer driver genes. Nevertheless, HBP1 expression is lower in several tumor types relative to matched normal tissues. Several micro-RNAs, such as miR-155, miR-17-92, and miR-29a, dampen HBP1 expression in cancer cells of various origins. The phosphatidylinositol-3 kinase (PI3K)/AKT pathway also inhibits HBP1 transcription by preventing FOXO binding to the HBP1 promoter. In addition, AKT directly phosphorylates HBP1, thereby inhibiting its transcriptional activity. Taken together, these findings place HBP1 at the center of a network of micro-RNAs and oncoproteins that control cell proliferation. In this review, we discuss our current understanding of HBP1 function in human physiology and diseases.





Similar content being viewed by others
References
Lesage F, Hugnot JP, Amri EZ, Grimaldi P, Barhanin J, Lazdunski M (1994) Expression cloning in K+ transport defective yeast and distribution of HBP1, a new putative HMG transcriptional regulator. Nucleic Acids Res 22:3685–3688
Tevosian SG, Shih HH, Mendelson KG, Sheppard KA, Paulson KE, Yee AS (1997) HBP1: a HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev 11:383–396
Lavender P, Vandel L, Bannister AJ, Kouzarides T (1997) The HMG-box transcription factor HBP1 is targeted by the pocket proteins and E1A. Oncogene 14:2721–2728
Song X, Gao X, Lu J, Liang H, Su P, Li Q, Pang Y (2018) High mobility group box transcription factor 1 (HBP1) from Lampetra japonica affects cell cycle regulation. Dev Growth Differ 60:146–157
Giese K, Amsterdam A, Grosschedl R (1991) DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev 5:2567–2578
Zhuma T, Tyrrell R, Sekkali B, Skavdis G, Saveliev A, Tolaini M, Roderick K, Norton T, Smerdon S, Sedgwick S, Festenstein R, Kioussis D (1999) Human HMG box transcription factor HBP1: a role in hCD2 LCR function. EMBO J 18:6396–6406
de Chiara C, Giannini C, Adinolfi S, de Boer J, Guida S, Ramos A, Jodice C, Kioussis D, Pastore A (2003) The AXH module: an independently folded domain common to ataxin-1 and HBP1. FEBS Lett 551:107–112
de Chiara C, Menon RP, Adinolfi S, de Boer J, Ktistaki E, Kelly G, Calder L, Kioussis D, Pastore A (2005) The AXH domain adopts alternative folds the solution structure of HBP1 AXH. Structure 13:743–753
Wang W, Pan K, Chen Y, Huang C, Zhang X (2012) The acetylation of transcription factor HBP1 by p300/CBP enhances p16INK4A expression. Nucleic Acids Res 40:981–995
Swanson KA, Knoepfler PS, Huang K, Kang RS, Cowley SM, Laherty CD, Eisenman RN, Radhakrishnan I (2004) HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat Struct Mol Biol 11:738–746
Bollaert E, Johanns M, Herinckx G, de Rocca Serra A, Vandewalle VA, Havelange V, Rider MH, Vertommen D, Demoulin JB (2018) HBP1 phosphorylation by AKT regulates its transcriptional activity and glioblastoma cell proliferation. Cell Signal 44:158–170
Zhang X, Kim J, Ruthazer R, McDevitt MA, Wazer DE, Paulson KE, Yee AS (2006) The HBP1 transcriptional repressor participates in RAS-induced premature senescence. Mol Cell Biol 26:8252–8266
Shih HH, Tevosian SG, Yee AS (1998) Regulation of differentiation by HBP1, a target of the retinoblastoma protein. Mol Cell Biol 18:4732–4743
Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, Yen HC, Elledge SJ (2011) Global identification of modular cullin-RING ligase substrates. Cell 147:459–474
Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteom 10(M111):013284
Lampert F, Stafa D, Goga A, Soste MV, Gilberto S, Olieric N, Picotti P, Stoffel M, Peter M (2018) The multi-subunit GID/CTLH E3 ubiquitin ligase promotes cell proliferation and targets the transcription factor Hbp1 for degradation. Elife 7:e35528
Xiu M, Kim J, Sampson E, Huang CY, Davis RJ, Paulson KE, Yee AS (2003) The transcriptional repressor HBP1 is a target of the p38 mitogen-activated protein kinase pathway in cell cycle regulation. Mol Cell Biol 23:8890–8901
Wang S, Cao Z, Xue J, Li H, Jiang W, Cheng Y, Li G, Zhang X (2017) A positive feedback loop between Pim-1 kinase and HBP1 transcription factor contributes to hydrogen peroxide-induced premature senescence and apoptosis. J Biol Chem 292:8207–8222
Amaravadi R, Thompson CB (2005) The survival kinases Akt and Pim as potential pharmacological targets. J Clin Invest 115:2618–2624
Pan K, Chen Y, Roth M, Wang W, Wang S, Yee AS, Zhang X (2013) HBP1-mediated transcriptional regulation of DNA methyltransferase 1 and its impact on cell senescence. Mol Cell Biol 33:887–903
Chen Y, Pan K, Wang P, Cao Z, Wang W, Wang S, Hu N, Xue J, Li H, Jiang W, Li G, Zhang X (2016) HBP1-mediated regulation of p21 protein through the Mdm2/p53 and TCF4/EZH2 pathways and its impact on cell senescence and tumorigenesis. J Biol Chem 291:12688–12705
Lemercier C, Duncliffe K, Boibessot I, Zhang H, Verdel A, Angelov D, Khochbin S (2000) Involvement of retinoblastoma protein and HBP1 in histone H1(0) gene expression. Mol Cell Biol 20:6627–6637
Escamilla-Powers JR, Daniel CJ, Farrell A, Taylor K, Zhang X, Byers S, Sears R (2010) The tumor suppressor protein HBP1 is a novel c-myc-binding protein that negatively regulates c-myc transcriptional activity. J Biol Chem 285:4847–4858
Berasi SP, Xiu M, Yee AS, Paulson KE (2004) HBP1 repression of the p47phox gene: cell cycle regulation via the NADPH oxidase. Mol Cell Biol 24:3011–3024
Li H, Wang W, Liu X, Paulson KE, Yee AS, Zhang X (2010) Transcriptional factor HBP1 targets P16(INK4A), upregulating its expression and consequently is involved in Ras-induced premature senescence. Oncogene 29:5083–5094
Sampson EM, Haque ZK, Ku MC, Tevosian SG, Albanese C, Pestell RG, Paulson KE, Yee AS (2001) Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. EMBO J 20:4500–4511
Elfert S, Weise A, Bruser K, Biniossek ML, Jagle S, Senghaas N, Hecht A (2013) Acetylation of human TCF4 (TCF7L2) proteins attenuates inhibition by the HBP1 repressor and induces a conformational change in the TCF4:DNA complex. PLoS One 8:e61867
Claeys S, Denecker G, Durinck K, Decaesteker B, Mus LM, Loontiens S, Vanhauwaert S, Althoff K, Wigerup C, Bexell D, Dolman E, Henrich KO, Wehrmann L, Westerhout EM, Demoulin JB, Kumps C, Van Maerken T, Laureys G, Van Neste C, De Wilde B, De Wever O, Westermann F, Versteeg R, Molenaar JJ, Pahlman S, Schulte JH, De Preter K, Speleman F (2018) ALK positively regulates MYCN activity through repression of HBP1 expression. Oncogene. https://doi.org/10.1038/s41388-018-0595-3
Zhang Y, Gao Y, Zhao L, Han L, Lu Y, Hou P, Shi X, Liu X, Tian B, Wang X, Huang B, Lu J (2013) Mitogen-activated protein kinase p38 and retinoblastoma protein signalling is required for DNA damage-mediated formation of senescence-associated heterochromatic foci in tumour cells. FEBS J 280:4625–4639
Lin KM, Zhao WG, Bhatnagar J, Zhao WD, Lu JP, Simko S, Schueneman A, Austin GE (2001) Cloning and expression of human HBP1, a high mobility group protein that enhances myeloperoxidase (MPO) promoter activity. Leukemia 15:601–612
Yao CJ, Works K, Romagnoli PA, Austin GE (2005) Effects of overexpression of HBP1 upon growth and differentiation of leukemic myeloid cells. Leukemia 19:1958–1968
Chan CY, Yu P, Chang FT, Chen ZH, Lee MF, Huang CY (2018) Transcription factor HMG box-containing protein 1 (HBP1) modulates mitotic clonal expansion (MCE) during adipocyte differentiation. J Cell Physiol 233:4205–4215
Borrelli S, Candi E, Hu B, Dolfini D, Ravo M, Grober OM, Weisz A, Dotto GP, Melino G, Vigano MA, Mantovani R (2010) The p63 target HBP1 is required for skin differentiation and stratification. Cell Death Differ 17:1896–1907
Paulson KE, Rieger-Christ K, McDevitt MA, Kuperwasser C, Kim J, Unanue VE, Zhang X, Hu M, Ruthazer R, Berasi SP, Huang CY, Giri D, Kaufman S, Dugan JM, Blum J, Netto G, Wazer DE, Summerhayes IC, Yee AS (2007) Alterations of the HBP1 transcriptional repressor are associated with invasive breast cancer. Cancer Res 67:6136–6145
Liang H, Fairman J, Claxton DF, Nowell PC, Green ED, Nagarajan L (1998) Molecular anatomy of chromosome 7q deletions in myeloid neoplasms: evidence for multiple critical loci. Proc Natl Acad Sci USA 95:3781–3785
Koike M, Tasaka T, Spira S, Tsuruoka N, Koeffler HP (1999) Allelotyping of acute myelogenous leukemia: loss of heterozygosity at 7q31.1 (D7S486) and q33-34 (D7S498, D7S505). Leuk Res 23:307–310
Driouch K, Briffod M, Bieche I, Champeme MH, Lidereau R (1998) Location of several putative genes possibly involved in human breast cancer progression. Cancer Res 58:2081–2086
Zenklusen JC, Thompson JC, Troncoso P, Kagan J, Conti CJ (1994) Loss of heterozygosity in human primary prostate carcinomas: a possible tumor suppressor gene at 7q31.1. Cancer Res 54:6370–6373
Zenklusen JC, Weitzel JN, Ball HG, Conti CJ (1995) Allelic loss at 7q31.1 in human primary ovarian carcinomas suggests the existence of a tumor suppressor gene. Oncogene 11:359–363
Zenklusen JC, Thompson JC, Klein-Szanto AJ, Conti CJ (1995) Frequent loss of heterozygosity in human primary squamous cell and colon carcinomas at 7q31.1: evidence for a broad range tumor suppressor gene. Cancer Res 55:1347–1350
Chen YC, Zhang XW, Niu XH, Xin DQ, Zhao WP, Na YQ, Mao ZB (2010) Macrophage migration inhibitory factor is a direct target of HBP1-mediated transcriptional repression that is overexpressed in prostate cancer. Oncogene 29:3067–3078
Tseng RC, Huang WR, Lin SF, Wu PC, Hsu HS, Wang YC (2014) HBP1 promoter methylation augments the oncogenic beta-catenin to correlate with prognosis in NSCLC. J Cell Mol Med 18:1752–1761
Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, Liu M, Zou Y, Weissman IL, Gu H (2010) microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med 207:475–489
Li H, Bian C, Liao L, Li J, Zhao RC (2011) miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat 126:565–575
Coomans de Brachene A, Bollaert E, Eijkelenboom A, de Rocca Serra A, van der Vos KE, Burgering BM, Coffer PJ, Essaghir A, Demoulin JB (2014) The expression of the tumour suppressor HBP1 is down-regulated by growth factors via the PI3K/PKB/FOXO pathway. Biochem J 460:25–34
Chen Y, Wang Y, Yu Y, Xu L, Zhang Y, Yu S, Li G, Zhang Z (2016) Transcription factor HBP1 enhances radiosensitivity by inducing apoptosis in prostate cancer cell lines. Anal Cell Pathol (Amst) 2016:7015659
Coomans de Brachene A, Demoulin JB (2016) FOXO transcription factors in cancer development and therapy. Cell Mol Life Sci 73:1159–1172
Chan CY, Huang SY, Sheu JJ, Roth MM, Chou IT, Lien CH, Lee MF, Huang CY (2017) Transcription factor HBP1 is a direct anti-cancer target of transcription factor FOXO1 in invasive oral cancer. Oncotarget 8:14537–14548
Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B, Ng PK, Jeong KJ, Cao S, Wang Z, Gao J, Gao Q, Wang F, Liu EM, Mularoni L, Rubio-Perez C, Nagarajan N, Cortes-Ciriano I, Zhou DC, Liang WW, Hess JM, Yellapantula VD, Tamborero D, Gonzalez-Perez A, Suphavilai C, Ko JY, Khurana E, Park PJ, Van Allen EM, Liang H, Group MCW, N Cancer Genome Atlas Research, Lawrence MS, Godzik A, Lopez-Bigas N, Stuart J, Wheeler D, Getz G, Chen K, Lazar AJ, Mills GB, Karchin R, Ding L (2018) Comprehensive characterization of cancer driver genes and mutations. Cell 173:371–385 e18
He S, Yang S, Niu M, Zhong Y, Dan G, Zhang Y, Ma H, Xiong W, Zhou M, Zhou Y, Xiang B, Li G, Shuai C, Peng S (2018) HMG-box transcription factor 1: a positive regulator of the G1/S transition through the Cyclin-CDK-CDKI molecular network in nasopharyngeal carcinoma. Cell Death Dis 9:100
Sanghvi VR, Mavrakis KJ, Van der Meulen J, Boice M, Wolfe AL, Carty M, Mohan P, Rondou P, Socci ND, Benoit Y, Taghon T, Van Vlierberghe P, Leslie CS, Speleman F, Wendel HG (2014) Characterization of a set of tumor suppressor microRNAs in T cell acute lymphoblastic leukemia. Sci Signal 7:ra111
Georgakilas AG, Martin OA, Bonner WM (2017) p21: a two-faced genome guardian. Trends Mol Med 23:310–319
Spiller CM, Wilhelm D, Jans DA, Bowles J, Koopman P (2017) Mice lacking Hbp1 function are viable and fertile. PLoS One 12:e0170576
Watanabe N, Kageyama R, Ohtsuka T (2015) Hbp1 regulates the timing of neuronal differentiation during cortical development by controlling cell cycle progression. Development 142:2278–2290
Shih HH, Xiu M, Berasi SP, Sampson EM, Leiter A, Paulson KE, Yee AS (2001) HMG box transcriptional repressor HBP1 maintains a proliferation barrier in differentiated liver tissue. Mol Cell Biol 21:5723–5732
Sekkali B, Szabat E, Ktistaki E, Tolaini M, Roderick K, Harker N, Patel A, Williams K, Norton T, Kioussis D (2005) Human high mobility group box transcription factor 1 affects thymocyte development and transgene variegation. J Immunol 175:5203–5212
Dong Z, Huang M, Liu Z, Xie P, Dong Y, Wu X, Qu Z, Shen B, Huang X, Zhang T, Li J, Liu J, Yanase T, Zhou C, Xu Y (2016) Focused screening of mitochondrial metabolism reveals a crucial role for a tumor suppressor Hbp1 in ovarian reserve. Cell Death Differ 23:1602–1614
Raine EV, Wreglesworth N, Dodd AW, Reynard LN, Loughlin J (2012) Gene expression analysis reveals HBP1 as a key target for the osteoarthritis susceptibility locus that maps to chromosome 7q22. Ann Rheum Dis 71:2020–2027
Martinez-Jacobo L, Cordova-Fletes C, Ortiz-Lopez R, Rivas F, Saucedo-Carrasco C, Rojas-Martinez A (2013) Delineation of a de novo 7q21.3q31.1 deletion by CGH-SNP arrays in a girl with multiple congenital anomalies including severe glaucoma. Mol Syndromol 4:285–291
van der Laan SW, Siemelink MA, Haitjema S, Foroughi Asl H, Perisic L, Mokry M, van Setten J, Malik R, Dichgans M, Worrall BB, MCotISG Consortium, Samani NJ, Schunkert H, Erdmann J, Hedin U, Paulsson-Berne G, Bjorkegrenn JLM, de Borst GJ, Asselbergs FW, den Ruijter FW, de Bakker PIW, Pasterkamp G (2018) Genetic susceptibility loci for cardiovascular disease and their impact on atherosclerotic plaques. Circ Genom Precis Med 11:e002115
Young RS, Weaver DD, Kukolich MK, Heerema NA, Palmer CG, Kawira EL, Bender HA (1984) Terminal and interstitial deletions of the long arm of chromosome 7: a review with five new cases. Am J Med Genet 17:437–450
Tzschach A, Menzel C, Erdogan F, Schubert M, Hoeltzenbein M, Barbi G, Petzenhauser C, Ropers HH, Ullmann R, Kalscheuer V (2007) Characterization of a 16 Mb interstitial chromosome 7q21 deletion by tiling path array CGH. Am J Med Genet A 143:333–337
Fagan K, Gill A, Henry R, Wilkinson I, Carey B (1989) A summary of 7q interstitial deletions and exclusion mapping of the gene for beta-glucuronidase. J Med Genet 26:619–625
Al-Hassnan ZN, Al-Bakheet A, Abu-Dheim N, Al-Younes B, Colak D, Kaya N (2011) A novel interstitial microdeletion of 7q22.1-7q22.3 detected by array comparative genomic hybridization. Am J Med Genet A 155A:3128–3131
Li Y, Choi PS, Casey SC, Dill DL, Felsher DW (2014) MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell 26:262–272
Kayali S, Giraud G, Morle F, Guyot B (2012) Spi-1, Fli-1 and Fli-3 (miR-17-92) oncogenes contribute to a single oncogenic network controlling cell proliferation in friend erythroleukemia. PLoS One 7:e46799
Wu H, Ng R, Chen X, Steer CJ, Song G (2016) MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut 65:1850–1860
Yan Z, Wang J, Wang C, Jiao Y, Qi W, Che S (2014) miR-96/HBP1/Wnt/beta-catenin regulatory circuitry promotes glioma growth. FEBS Lett 588:3038–3046
Wan YC, Li T, Han YD, Zhang HY, Lin H, Zhang B (2016) MicroRNA-155 enhances the activation of Wnt/beta-catenin signaling in colorectal carcinoma by suppressing HMG-box transcription factor 1. Mol Med Rep 13:2221–2228
Sun X, Geng X, Zhang J, Zhao H, Liu Y (2015) miR-155 promotes the growth of osteosarcoma in a HBP1-dependent mechanism. Mol Cell Biochem 403:139–147
Yan Z, Che S, Wang J, Jiao Y, Wang C, Meng Q (2015) miR-155 contributes to the progression of glioma by enhancing Wnt/beta-catenin pathway. Tumour Biol 36:5323–5331
Yang Z, Wu L, Zhu X, Xu J, Jin R, Li G, Wu F (2013) MiR-29a modulates the angiogenic properties of human endothelial cells. Biochem Biophys Res Commun 434:143–149
Tian FJ, An LN, Wang GK, Zhu JQ, Li Q, Zhang YY, Zeng A, Zou J, Zhu RF, Han XS, Shen N, Yang HT, Zhao XX, Huang S, Qin YW, Jing Q (2014) Elevated microRNA-155 promotes foam cell formation by targeting HBP1 in atherogenesis. Cardiovasc Res 103:100–110
Chen H, Li X, Liu S, Gu L, Zhou X (2017) MircroRNA-19a promotes vascular inflammation and foam cell formation by targeting HBP-1 in atherogenesis. Sci Rep 7:12089
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bollaert, E., de Rocca Serra, A. & Demoulin, JB. The HMG box transcription factor HBP1: a cell cycle inhibitor at the crossroads of cancer signaling pathways. Cell. Mol. Life Sci. 76, 1529–1539 (2019). https://doi.org/10.1007/s00018-019-03012-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03012-9