Abstract
B cell leukaemia is one of the most frequent malignancies in the paediatric population, but also affects a significant proportion of adults in developed countries. The majority of infant and paediatric cases initiate the process of leukaemogenesis during foetal development (in utero) through the formation of a chromosomal translocation or the acquisition/deletion of genetic material (hyperdiploidy or hypodiploidy, respectively). This first genetic insult is the major determinant for the prognosis and therapeutic outcome of patients. B cell leukaemia in adults displays similar molecular features as its paediatric counterpart. However, since this disease is highly represented in the infant and paediatric population, this review will focus on this demographic group and summarise the biological, clinical and epidemiological knowledge on B cell acute lymphoblastic leukaemia of four well characterised subtypes: t(4;11) MLL-AF4, t(12;21) ETV6-RUNX1, t(1;19) E2A-PBX1 and t(9;22) BCR-ABL1.
Similar content being viewed by others
Introduction
Cancer is the top fatal disease in the paediatric population and now shares the first place in adults with heart disease [1]. It is a multifactorial disease that is influenced by genetics (e.g. chromosomal translocations, point mutations and single nucleotide polymorphisms), lifestyle (e.g. tobacco consumption and obesity), exposure to environmental factors (e.g. sunlight, radiation and infectious agents) and aging [2,3,4]. Over a lifetime, humans acquire a combination of genetic insults. Depending on the genetic insults and the cell-of-origin, this translates into the development of a sporadic cancer (~90% of cancers).
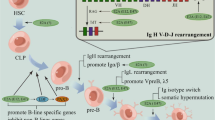
The scientific community has noted a continued increase of sporadic cancers in the paediatric population since the 1970s. This increase is strikingly restricted to developed countries, suggesting that our lifestyle plays a significant role in cancer development. Epidemiological factors that have been linked to an increased risk of developing paediatric B cell acute lymphoblastic leukaemia (B-ALL) include pesticides [5], magnetic exposure [6] and an overstimulation of the immune response during the first years of life (population-mixing and delayed-infection hypothesis) [7,8,9]. B-ALL accounts for 26% of cancers diagnosed in children aged 0–14 years old [10], making it one of the most prevalent cancers in the paediatric population. This disease leads to the differentiation arrest of B lymphoid cells at an immature stage (progenitor or precursor), followed by an uncontrolled proliferation and accumulation of leukaemia blasts in the bone marrow. These pro/pre-B leukaemia blasts will migrate from the bone marrow to the peripheral circulation to infiltrate the spleen, liver, thymus, lymph nodes and central nervous system. This hijack of the haematopoietic system by leukaemia blasts also compromises the production of other essential blood cell types for oxygen transport (erythrocytes) or coagulation (platelets). This leads to major insufficiencies in the patient’s immunological and physiological system, which ultimately results in death. Early signs of the disease include fatigue, loss of appetite, bone pain, swelling in the abdomen (due to splenomegaly and hepatomegaly) and swelling of the lymph nodes. Acute leukaemia has a rapid progression (weeks) compared to chronic leukaemia (months).
Therapies for B-ALL have greatly improved since the 1970s when the survival rate was only 10% [11]. Today, at least 90% of paediatric patients and 40% of adult patients will recover from this disease [12]. This treatment success is explained by breakthroughs in understanding the molecular mechanisms of leukaemogenesis, leading to the development of efficient and specific therapies and the use of stem-cell transplantation. The biology of B-ALL is determined mainly by the first genetic hit, which is usually a change in chromosome numbers (hyperdiploidy or hypodiploidy) or a chromosomal translocation (e.g. t(4;11) MLL-AF4 or t(12;21) ETV6-RUNX1) (Table 1). In paediatric patients, this genetic insult can have an in utero origin that can be traced back to birth by analysing the Guthrie cards which contain newborn blood spots [13]. Increasing evidence suggests that additional genetic events are required for leukaemia development, which will be described in each section. Furthermore, patients at relapse show deletions at specific loci (e.g. CDKN2A/B, ETV6, IKZF1, NR3C1) [14]. Some patients also acquire mutations in NT5C2, leading to drug-resistance [15], and in CREBBP which impairs the acetylation and transcriptional regulation of CREBBP-target genes [16].
This review will explore the biological mechanisms that are affected by four chromosomal translocations: t(4;11) MLL-AF4, t(12;21) ETV6-RUNX1, t(1;19) E2A-PBX1 and t(9;22) BCR-ABL1. The first three are associated with perturbations in the gene expression profile, whereas t(9;22) BCR-ABL1 hijacks signalling pathways. Since the members of each translocation are involved in normal haematopoiesis, we will also describe their role in a physiological situation. We have chosen to focus on these four subclasses as they cover around 40% of B-ALL patients in children and adults (and 80% in infants) and they have been well studied in patients and animal models. However, for completeness’ sake, we will also briefly mention other subtypes: including hyperdiploidy, hypodiploidy, trisomy 4 and 10 and intraamplification of chromosome 21. In addition, the therapeutic avenues currently available in the clinic will be discussed and potential new agents highlighted that are in clinical trials (Table 2).
The fast and the furious: t(4;11)(q21;q23) MLL-AF4
Clinical features of MLL-AF4+ patients
The rearrangement of the MLL gene (11q23) is a common genetic event in haematological malignancies [17]. It is present in around 10% of ALL and 5% of acute myeloid leukaemia (AML). There are more than 80 genes that can form chromosomal translocations with the MLL gene in leukaemia, with AF4, AF9 and ENL amongst the most common. MLL-rearrangements are found in both paediatric and adult ALL, but the prognosis is poorer in childhood ALL regardless of the age at diagnosis [18]. Around 50% of patients diagnosed with pro-B ALL aged 6 months or less harbour the t(4;11) MLL-AF4 chromosomal rearrangement [19]. The frequency of t(4;11) MLL-AF4 ALL decreases in older infants (10–20%) and drops to just 2% in children. In adult B-ALL, it accounts for around 10% of cases.
t(4;11)+ leukaemia is associated with an increased leukocyte count due to an accumulation of leukaemia blasts with a phenotype similar to progenitor B cells (negative for CD10) [18]. The leukaemia blasts often co-express haematopoietic progenitor markers (CD34), B lymphoid markers (CD19, CD22, CD79a, Tdt), myeloid markers (CD15, CD65) and components of the leukocyte antigen complex (HLA-DR) [20]. They are negative for CD24, IgM, CD13 and CD33. A significant proportion of adult and paediatric patients express the neural glial marker NG2 on blast cells [21, 22], which is also detected in t(12;21) ETV6-RUNX1 B-ALL and t(9;11) MLL-AF9 AML [23]. Interestingly, the NG2 antigen is not expressed in haematopoietic stem cells derived from adult bone marrow or neonatal cord blood, but is expressed in more mature haematopoietic progenitor cells [23, 24].
An interesting feature of t(4;11)+ infant patients is the expression of MLL-AF4 in a subset of bone marrow mesenchymal stem cells (MSC) [25]. This was not observed in other types of B-ALL and seems to be unique to this group of patients. It suggests that the initial translocation occurred in a precursor for both the haematopoietic and the stromal compartment. To investigate the impact of MLL-AF4 expression in early development, Bueno et al. induced the expression of this fusion gene through a lentiviral vector in human embryonic stem cells (hESC) with expression levels similar to those found in patient-derived cell lines [26]. MLL-AF4 induced an upregulation of HOX gene expression and enhanced the formation of haemogenic precursor cells (CD45−CD31+CD34+). This was accompanied by a diminished haematopoietic differentiation and an increased endothelial cell fate. Therefore, MLL-AF4 does not seem to favour haematopoietic commitment during the mesodermal to haematopoietic transition; however, MLL-AF4 expression in the stromal compartment may increase niche support for the cell-of-origin and the leukaemia blasts.
MLL, a regulator of transcription essential for embryonic and adult haematopoiesis
MLL, also known as KMT2A, is an H3K4 methyltransferase that catalyses histone H3 Lys4 methylation through its SET domain. This epigenetic mark is associated with actively transcribed promoters and open chromatin. MLL directly interacts with the nuclear protein Menin (MEN1), which is essential for MLL-mediated transcription and for MLL-AF4-mediated transformation [27, 28]. Accordingly, MEN1 inhibitors were shown to be detrimental to oncogenic transformation by MLL-fusion genes, independent of the MLL partner gene [29, 30]. Inhibitors of WDR5 and LEDGF, two partners of MLL, can also disrupt MLL activity and the oncogenic potential of MLL-AF4 [31,32,33].
MLL plays a vital role during embryonic development by regulating HOX gene expression, which is crucial for the specification of segment identity [34]. Mll haploinsufficiency in mice leads to major disorders in the cervical, lumbar and thoracic regions. Hence, Mll is critical for pattern formation and proper development of the embryo. A complete knock-out of Mll in mice leads to death at embryonic day (E)10.5 because of dysplasia in the branchial arch and aberrant segment boundaries of spinal ganglia and somites [35]. E10.5 is also the developmental time-point when the first definitive haematopoietic stem cells (HSCs) emerge in the aorta-gonads-mesonephros (AGM) region in a process that depends on Runx1, a transcription factor linked to pre-B ALL ([36, 37] and see below). Subsequent work from the Korsmeyer group has shown that Mll is important for maintaining haematopoietic potential throughout embryonic development. Mll is essential for the haematopoietic colony-forming potential and proliferation of haematopoietic progenitors in the E10.5 yolk sac [38], the tissue in which haematopoietic cells are first detected [39]. Mll continues to have a role in maintaining the haematopoietic potential at later stages in the E12.5 foetal liver and yolk sac [40]. Furthermore, Mll-deficient embryonic stem (ES) cells cannot reconstitute haematopoietic organs in chimeric animals, leading to a lack of haematopoietic potential in the foetal liver [41]. Importantly, Mll-null AGMs are devoid of HSCs [41]. Mll also has a cell-intrinsic role in the self-renewal of HSCs from the foetal liver and adult bone marrow since its deletion leads to an impaired engraftment in competitive transplants [42]. Hence, Mll is a gene that is essential to maintain the haematopoietic potential during both embryonic and adult development.
AF4, a mediator of RNA elongation essential to lymphopoiesis
AF4 is a member of the AF4/FMR2 family that contains four members: AFF1/AF4, AFF2/FMR2, AFF3/LAF4 and AFF4/AF5q31. Most members, with the exception of AFF2/FMR2, can translocate with the MLL gene and participate in the development of ALL or AML. AF4 is part of the AEP complex, which includes other members of the AF4/FMR2 family (AF5Q31), the ENL family (ENL and AF9) and the p-TEFB elongation factor. The AEP complex is important for releasing the paused RNA polymerase II, which initiates RNA elongation. As mentioned previously, MLL can fuse to more than 80 different partner genes in haematological malignancies, most of which are members of the AEP complex. Some members of this family (AFF2/FMR2, AFF3/LAF4 and AFF4/AF5q31) also localise to nuclear speckles which are structures containing pre-mRNA splicing factors [43]. Those structures contain the regulatory subunit cyclin T1 and the catalytic domain CDK9, which together form the p-TEFB elongation factor. P-TEFB can be inactivated by flavopiridol [44], which has completed its phase I clinical trial for recurrent B-ALL in adults (NCT00278330). Hence, some members of the AF4/FMR2 family can also participate in the splicing of messenger RNA, and this process could be tightly associated with RNA elongation. However, AF4 does not localise to nuclear speckles, so it is unlikely that the MLL-AF4 fusion gene can deregulate this pathway.
Af4 is ubiquitously expressed, but its level of expression is higher in the lymphoid compartment and placenta [45, 46]. Af4 −/− knockout mice show a severe defect in lymphopoiesis compared with Af4 +/− mice, as evidenced by reduced numbers of B and T cells in the main adult haematopoietic sites such as the bone marrow, spleen and thymus [47]. AF4 can also promote the expression of CD133, a cell surface marker of hematopoietic and cancer stem cells [48]. The immortalisation of myeloid progenitors by the MLL-AF4 fusion gene requires the AF4-binding platform (pSER domain) as shown in colony replating assays [49]. AF4 is also important for recruiting selectivity factor 1 (SL1), which is a specific pSER domain binder, and this ensures the loading of TBP to the TATA box [50]. This study provides new evidence for a transactivation role of AF4 in the leukaemogenesis process. The N-terminal part of AF4 can bind the pTEFb complex, but also recruit TFIIH and MEN1 [51]. This is interesting since the AF4-MLL reciprocal fusion gene has also been implicated in B-ALL development. This will be discussed later in this section.
The biology of t(4;11) MLL-AF4 infant leukaemia
Cancer development is a disease that is normally associated with the acquisition of an array of mutations throughout a lifetime. Paediatric ALL, however, has one of the lowest mutation rates, which is estimated at approximately 0.2–0.4 mutations per MegaBase [52]. Since this disease is usually initiated in utero at a developmental stage where the chromatin is more open and accessible than in adults [53], it is possible that all the factors required by MLL-AF4 to initiate disease are already active. Whole-genome, exome and targeted DNA sequencing studies in t(4;11) infant patients have confirmed the absence of cooperating mutations in this sub-type of leukaemia [54]. A recent study using ultra deep sequencing of 36 paediatric t(4;11) MLL-AF4 ALL found that 63.9% of them had a mutation in NRAS or KRAS that could lead to its hyperactivity [55]; however, experimental evidence suggests that these may influence the timing of disease onset and malignant cell migration, but are not required for disease initiation [56, 57], as further discussed below. It is thus clear that t(4;11)+ pro-B ALL has a lower mutation rate than other subtypes of B cell leukaemia, suggesting that MLL-AF4 alone is sufficient to initiate disease.
Several attempts have been made to generate a mouse model for MLL-AF4+ leukaemia. First, in 2006, Chen et al. developed a constitutive knock-in mouse, in which human AF4 was inserted into the mouse Mll locus [58]. They obtained some heterozygous Mll-AF4 knock-in animals, despite noticeable embryonic lethality. These were smaller at birth compared to their wild-type littermates, presented with a higher white blood cell count in the peripheral blood, increased levels of neutrophils and a bias toward the B lymphoid lineage [58]. Cells from Mll-AF4 knock-in mice produced more pro-B cell colonies in methylcellulose assays compared with their wild-type littermates, and could serially replate for three rounds. This suggested an increased self-renewal when Mll-AF4 is expressed and an immortalisation of the pro-B cells. However, when those cells were placed in liquid culture, they could not survive for more than two months, implying that the transformation process was not completed. The Mll-AF4 knock-in mice did develop haematological malignancies (lymphoid and myeloid hyperplasia, B cell lymphoma and myeloid malignancy) with a median time of 570 days. Therefore, MLL-AF4 on its own can lead to a haematological malignancy, but this model did not recapitulate the human disease.
Because of the high embryonic lethality rate observed in constitutive Mll-AF4 knock-in mice, Metzler et al. developed a conditional knock-in model based on the invertor technology and the use of specific Cre recombinases [59]. Using Cre recombinase expressed under the control of specific lymphocyte promoters, they successfully generated mice that expressed Mll-AF4 in B lymphocytes (CD19-Cre), T lymphocytes (Lck-Cre) or both (Rag-Cre). In all cases, the mice developed large diffuse B cell lymphomas with a latency of 317–475 days, which was shorter than the previous Mll-AF4 knock-in mice. Even though the phenotype of the disease was still dissimilar to the human counterpart, it revealed a bias of MLL-AF4-driven disease towards the B lineage even when targeted to T cells.
In 2008, Krivtsov et al. published their work with a new conditional knock-in mouse model of Mll-AF4, which used the Mx1-Cre system to activate the expression of Mll-AF4 postnatally after an induced interferon response [60]. Following multiple intraperitoneal injections of polyinosinic/polycytidylic acid (polyI:C) to initiate the expression of Mll-AF4, 36% of the mice developed a haematological malignancy with a latency of 152 days and a disease phenotype that was similar to pre-B ALL, while 27% developed AML. It thus appeared that all of these mouse models were missing a crucial element required for the development of a disease more representative of the human leukaemia. One of the hypotheses was that a human physiological context was required. With the emergence of humanized mice that have a compromised immune system and can, therefore, tolerate human haematopoietic cell transplants (as reviewed in Ref. [61]), it is possible to use human haematopoietic cells to enhance our understanding of pro-B ALL.
Thus, Montes et al. isolated human cord blood CD34+ haematopoietic stem and progenitor cells and transduced them with a lentiviral vector containing the human MLL-AF4 cDNA [62]. The authors found an increased engraftment of human cord blood cells in NSG mice when MLL-AF4 was expressed. This was accompanied by an increase in HOXA9 expression. The clonogenic potential of the cells was slightly enhanced in myeloid-forming conditions, but the mice did not develop any haematological malignancies. This study suggested that human cord blood cells might represent a stage of development that is too late to recapitulate the leukaemogenesis happening in patients. In 2016, Lin et al. used an alternative approach [63] and created a retroviral construct that contains the human MLL and the mouse Af4 gene parts. Compared to the human MLL-AF4 construct, their hybrid produced a higher retroviral titer. They transduced the lineage negative fraction (Lin-) of mouse adult bone marrow, which contains all of the haematopoietic stem and progenitor cells, and cultured these in myeloid or lymphoid conditions. Following transplantation of the cultured cells, the recipients developed AML, irrespective of the culture conditions. This may at least partly be due to the Lin− fraction being mostly enriched in mature myeloid progenitors such as GMP and CMP, which would be preferentially targeted. Interestingly, when they transduced human CD34+ cells derived from cord blood and transplanted these into NSG recipients, the mice developed pro-B ALL. The authors propose that the development of pro-B ALL requires a human haematopoietic context. It does not, however, seem to require a prehaematopoietic or hemangioblast precursor, or additional events like the reciprocal fusion gene AF4-MLL.
Infant t(4;11) MLL-AF4 pro-B ALL has an in utero origin [13]. Therefore, to understand the first stages of the disease, it is important to initiate the expression of MLL-AF4 in embryonic haematopoietic cells which have unique and different properties than adult haematopoietic cells (as reviewed in Ref. [64]). Barrett and Malouf et al., therefore, used the conditional Mll-AF4 invertor mice developed by Terry Rabbitts and two different Cre lines to target the expression of Mll-AF4 to foetal haematopoietic cells: Vav-Cre (definitive haematopoietic cells) and Vec-Cre (haemogenic endothelium) [65]. The experimental set-up allowed targeting of all the haematopoietic cells formed during the definitive wave of haematopoiesis, but also the haemogenic endothelium that gives rise to HSCs in the AGM (E10.5 of development), lymphoid progenitors (E9.5) and erythro-myeloid progenitors (E8.5) in the yolk sac [66, 67]. The effect of Mll-AF4 on emerging blood cells was then analysed throughout haematopoietic development in the embryo. Blood cells formed in the AGM and placenta (E11-E12) showed a slight lymphoid bias in colony assays, but their engraftment was similar to control cells, suggesting that the transformation process is not initiated at those early stages of definitive haematopoiesis. At later stages in the foetal liver (E12–E14), when haematopoiesis has established itself, the engraftment potential of foetal liver cells was significantly enhanced upon Mll-AF4 expression and the B lymphoid bias became stronger in colony-forming assays. This was also accompanied by a higher production of myeloid colonies. Most mice developed B cell lymphomas with a latency of 437 days (Vec-Cre) or 556 days (Vav-Cre). This study also shed light on the cell-of-origin of infant pro-B ALL, which will be discussed in the next section.
The lymphoid-primed multipotent progenitor (LMPP) as a potential cell-of-origin
Previous studies have suggested that leukaemia originates in a haematopoietic progenitor that has a lineage-restricted differentiation potential, but has retained stem cell-like features. Those stem-cell properties confer higher self-renewal and quiescence, which makes them resistant to chemotherapeutic agents that can target proliferative leukaemia blasts [68, 69]. Over the past few years, studies have shed light on the cell-of-origin, but the debate is still open, especially for B-ALL.
In one study, Cox et al. [70] found that the human CD34+CD19− and CD34+CD10− fractions derived from the bone marrow of B cell leukaemia patients aged 1–16 years old could engraft immunocompromised NOD/SCID mice and lead to the development of a leukaemia with a phenotype similar to patients. Long-term cultures of unsorted bone marrow became strongly enriched for CD34+CD19− and CD34+CD10− fractions with time, with a concomitant loss of the CD19+ and CD10+ fraction, which also correlated with their lack of engraftment in xenotransplantation. Therefore, they concluded that the target cells for leukaemia initiation in B-ALL are immature haematopoietic progenitors that have not committed to the B cell lineage.
Le Viseur et al. sorted leukaemia cells from B-ALL patients according to the expression of CD34 and CD19, followed by xenotransplantation [71]. CD34+CD19−, CD34+CD19+ and CD34−CD19+ fractions all engrafted robustly and could initiate leukaemia development with a phenotype similar to what was observed in patients and a mean latency of 11.7 weeks.
Aoki et al. reported that the CD34+CD38−CD19−CD33− bone marrow fraction of infant t(4;11) MLL-AF4 pro-B ALL patients contained normal haematopoietic stem and progenitor cells since they gave rise to normal myelopoiesis and lymphopoiesis in xenotransplantation [72]. The injected cells did not initiate leukaemia or expressed MLL-AF4. However, xenotransplantation of CD34+CD38+CD19+ or CD34−CD19+ cells did lead to leukaemia development, and those cells expressed MLL-AF4. The expression of CD19 seems important for the leukaemia potential in xenotransplantation, but as mentioned previously, CD19− cells derived from patients can engraft and initiate leukaemia development in other studies [70, 71]. At a similar time, Bardini et al. found that the expression of CD34 and NG2 cannot differentiate leukaemia-initiating cells from non leukaemia-initiating cells [73]. They also found some Ig/TCR rearrangement in the xenografted cells, suggesting that the founder clone could be a haematopoietic progenitor that has already undergone some rearrangement (CD19+). There is also a possibility that the founder clone could be a cell that has no mature antigen receptor rearrangement (e.g. IGH, IGK-Kde, TCRD, TCRG) rearrangement (CD19−), which was observed in leukaemia cells from t(4;11) pro-B ALL patients [74]. The question of whether transformation begins before the commitment to the lymphoid lineage in patients, therefore, requires further investigation.
Using an inducible mouse model in which Mll-AF4 was targeted to the first definitive haematopoietic cells during development, Barrett and Malouf et al. found an increased B lymphoid output in the E12–E14 foetal liver during embryonic development compared to the control littermates [65]. They isolated two fractions: HSC/MPP (Lineage− Sca1+ ckit+ Flt3− IL7R−; stem cell and immature progenitors) and LMPP (Lineage− Sca1+ ckit+ Flt3+) and observed a notable increase in the formation of B cell colonies from both populations upon expression of Mll-AF4. These colonies were bigger in size compared to colonies formed by control cells and contained mostly pro-B cells (AA4.1−ckit+B220+CD19+CD43+CD24+IgM−) that could form colonies in replating assays. The clonogenic potential of the LMPP was around fourfold higher compared to the HSC/MPP fraction. All together, these results confirm that leukaemogenesis is initiated in utero and that Mll-AF4 confers stem-cell like properties to embryonic haematopoietic progenitors. This study also suggests that the foetal liver LMPP could be the cell-of-origin of t(4;11) MLL-AF4 leukaemia [65].
MLL-AF4 is a positive regulator of clonogenicity and survival
To gain further insights into the role of MLL-AF4 in leukaemia, scientists have used various human and mouse cell lines to assess its effect on survival, proliferation and clonogenic potential. Caslini et al. developed a tetracycline-inducible system of MLL-AF4 expression in U937 human monocyte cells and observed that an increased expression of MLL-AF4 leads to an inhibition of proliferation [75]. This phenotype was similar to the overexpression of MLL-AF9 [75]. By contrast, Thomas et al., using siRNA technology to inhibit the expression of MLL-AF4 in SEM and RS4;11 cells, observed that the inhibition of MLL-AF4 had a negative effect on the clonogenic potential and proliferation of leukaemia cells [76]. This was accompanied by a decreased engraftment in SCID mice and an upregulation of CD133, suggesting that the cells had started to differentiate and were losing their self-renewal potential. These seemingly contrasting results suggest that either the cell context can impact on the transformation process mediated by MLL-AF4 or maintaining the right level of MLL-AF4 expression is crucial, with knockdown and overexpression both having a negative impact on cell proliferation. The former interpretation is supported by recent work from the Mulloy group who revealed an impaired ability of MLL-Af4 to transform myeloid cells [77]. This lymphoid preference is driven by the Af4 partner and requires a lymphoid cellular context. The expression of MLL-AF4 can also be modulated by specific microRNAs that can recognize the 3′UTR of AF4, notably miR-143 [78]. The miR-143 locus is highly methylated in MLL-AF4+ leukaemia cell lines, leading to its downregulation and MLL-AF4 escaping microRNA-mediated regulation. Restoring the expression of miR-143 in MLL-AF4+ leukaemia cells leads to an increase in apoptosis and a decrease in cell proliferation. Hence, microRNAs can influence leukaemogenesis and bring an additional layer of complexity to our understanding of pro-B ALL.
Contributors to t(4;11) MLL-AF4 leukaemia
All the mice that express MLL-AF4 do develop a haematological malignancy; however, they do not recapitulate the human disease that has a fast progression and produces leukaemia blasts with a pro-B cell phenotype. These models suggest that additional events or factors are necessary, including changes in gene expression which may not be present in the murine context.
FLT3 activity in infant leukaemia
The activity of FLT3 is increased in t(4;11) MLL-AF4 pro-B ALL and other cytogenetic groups including t(12;21) ETV6-RUNX1 and t(9;22) BCR-ABL1 [79]. This can result from mutations in the tyrosine kinase domain (TKD) or a tandem duplication of the gene, but is usually associated with an increased expression of FLT3, commonly as a result of an upregulated HOXA9/MEIS1 program [80]. The prognosis of t(4;11) MLL-AF4 leukaemia can be predicted with FLT3 expression. High expression of FLT3 is associated with a bad prognosis, a shorter overall survival and a shorter disease-free survival, while low expression of FLT3 is associated with a better prognosis. To model the effect of FLT3, Montes et al. transduced human CD34+ cord blood cells with MLL-AF4 and either an overactive FLT3 (FLT3-TKD) or a wild-type FLT3 [81]. Using xenotransplantation, they assessed the development of leukaemia upon activation of FLT3. Even though the mice did not develop leukaemia, they observed a transient expansion of CD34+ cord blood cells expressing MLL-AF4 upon FLT3 activation.
AF4-MLL: the reciprocal fusion gene
The reciprocal fusion gene AF4-MLL is present in approximately 80% of patients diagnosed with t(4;11) MLL-AF4 leukaemia [82]. Similar to MLL-AF4, it contains cleavage sites for Threonine-Aspartase 1 (Taspase 1) which are important for its activation and its resistance to proteosomal degradation. AF4-MLL can associate with proteins that modulate the activity of RNA polymerase II promoters (e.g. P-TEFB kinase) and alter the histone methylation signature by recruiting H3 arginine and lysine methyltransferases, such as CARM1 and DOT1L, respectively, leading to an increased H3 methylation signature [83]. Therefore, both MLL-AF4 and AF4-MLL can increase H3K79, H3R2, H3R17 and H3R26 methylation marks. Additionally, AF4-MLL can increase the H3K4 methylation mark and extend the activation of gene expression. AF4-MLL can also interact with the C-terminal portion of AF4, suggesting that MLL-AF4 can be recruited to the same transcriptional complex. Neither MLL-AF4 nor AF4-MLL seem to affect the efficiency of DNA damage repair [84]. MLL-AF4 and AF4-MLL can modulate the transcription of the ALOX5 gene through the activation of transcription and elongation, respectively [85]. The ALOX5 gene encodes the 5-lipoxygenase enzyme, which is essential for the formation of pro-inflammatory leukotrienes. These metabolites of arachidonic acid can activate the immune response and help to establish a tumour microenvironment by acting both on cancer cells and stromal cells (as reviewed in Ref. [86]). They have also been linked to proliferation, as an increased production of leukotriene B4 in B cell chronic lymphocytic leukaemia was shown to enhance DNA synthesis [87]. Interestingly, it was shown that AF4-MLL can lead to leukaemia, even in the absence of MLL-AF4 [88]. Therefore, AF4-MLL appears to be important for leukaemia development, but its absence in 20% of patients implies it is not always essential.
NRAS and KRAS
The mutation of NRAS and KRAS in t(4;11) MLL-AF4 pro-B ALL has been reported in the literature despite the low mutation burden in this subtype of leukaemia [89]. MLL-AF4 can also activate the expression of the ELK-1 transcription factor, which is a downstream effector of KRAS activity [90]. This prompted scientists to study its role in leukaemogenesis. Crossing a transgenic MLL-AF4 mouse line with a transgenic Kras mouse line [57] resulted in the mice developing B cell lymphoma or leukaemia characterised by the accumulation of B220+CD43+CD19+ blasts, splenomegaly and infiltration of malignant cells in the lungs. The latency was significantly reduced by the presence of Kras mutations. Prieto et al. used xenotransplantation to assess the contribution of KRAS activation to the transformation of CD34+ cord blood cells expressing MLL-AF4 [56]. They found that the expression of KRAS favoured the migration of cells to extramedullary organs, but the mice did not develop leukaemia. A non-essential role for RAS signalling is also supported by the observation made in a recent large-scale sequencing study in which activating mutations in the RAS pathway were found to be mostly sub-clonal and often lost at relapse [54].
Transcriptional deregulation as a hallmark of MLL-rearranged leukaemia
The expression of MLL-AF4 is associated with an ectopic H3K79 methylation profile that is crucial for the transformation process in infant pro-B leukaemia and requires the aberrant recruitment of the DOT1L enzyme [60, 91]. Importantly, this is now explored as a potential therapeutic target [91, 92] with DOT1L inhibitors being currently tested in clinical trials. In addition, MLL-rearranged leukaemias have the highest DNA methylation index among all types of leukaemias [93].
The epigenetic abnormalities result in the upregulation in leukaemia blasts of an array of genes which are normally expressed in haematopoietic stem cells, such as HOXA9, MEIS1, HMGA2 and RUNX1 [37, 94,95,96]. Interestingly, RUNX1 was not only determined to be a direct target of MLL-AF4, but the RUNX1 protein was also found to form a complex with the reciprocal AF4-MLL fusion protein [97]. Around 50% of infant t(4;11) MLL-AF4 patients show an upregulation of HOX genes, which in combination with MEIS1, participate in the immortalisation process mediated by MLL-rearranged genes.
The HMG/high mobility group family of transcription factors (HMGA1 and HMGA2) are ubiquitously expressed during embryonic development, but almost undetectable in adult tissues. They are chromatin-remodelling factors that bind AT-rich regions of the minor groove of DNA. HMGA2 expression is upregulated in MLL-AF4 leukaemia cell lines which leads to an increased cell proliferation that is dependent on the hypermethylation of the miR-let-7b promoter [98]. This MLL-AF4/let-7b/HMGA2 axis is sensitive to the demethylating agent 5-azacytidine. Studies have suggested that MLL-AF4 can modulate the expression of BCL-2 target genes, which are members of the intrinsic mitochondrial apoptosis pathway [97, 99, 100]. In fact, MLL-AF4 can increase the expression of BCL-2 and MCL-1 and repress the transcription of BIM through binding of the promoters [28]. Accordingly, the inhibition of MLL-AF4 in the t(4;11)+ SEM cell line led to a decrease in H3K79me2/3 methylation at the BCL-2 locus and a decrease in H3K27Ac acetylation mark at the 3′ end [28, 100]. The expression of BCL-2 resulted in resistance to apoptosis, which could be reverted using a specific BCL-2-selective inhibitor ABT-199. Inhibition of MLL-AF4 also led to reduced binding of the PRC1 protein CBX8 to the BIM locus. Similar to the effects of MLL-AF4 inhibition, treatment of SEM cells with the DOT1L inhibitor EPZ5676 caused a loss of the histone marks and a decreased expression of BCL-2 and MCL-1, but not BIM.
In 2011, Dawson et al. investigated the effect of an inhibitor of the BET family I-BET151 on MLL-rearranged leukaemia [101]. I-BET151 works by removing the BRD3/4, PAFc and SEC components from chromatin. They found that this inhibitor had a strong effect on the survival of MLL-rearranged leukaemia cells derived from patients, and this was partly due to the inhibition of transcription of BCL-2, the oncogene C-MYC and CDK6, the latter of which is required for the exit of haematopoietic stem cells from quiescence [102]. At the moment, phase I clinical trials are underway using I-BET inhibitors (NCT01943851) [103] and inhibitors of DOT1L (NCT01684150, NCT02141828) [104]. The inhibitors for menin, LEDGF and WDR5 are still in their preclinical phases [105]. The biological features and therapeutic avenues of t(4;11) MLL-AF4 pro-B ALL are highlighted in Fig. 1.
Biology of t(4;11) MLL-AF4 pro-B acute lymphoblastic leukaemia. The MLL-AF4 fusion gene and AF4-MLL reciprocal fusion gene are shown with their main interaction partners. Both induce a deregulated epigenetic signature leading to an upregulation of stem-cell signature genes, positive regulators of cell division and inhibitors of apoptosis. The DOT1L inhibitor (EPZ5676), I-BET inhibitor (I-BET151), p-TEFB inhibitor (flavopiridol), WDR5 inhibitor (MM-401) and LEDGF inhibitor (CP65 small peptide) are potential therapeutic agents that target various members of the MLL-AF4 complex and its regulated genes
The usual suspect: t(12;21) ETV6-RUNX1
Clinical features of ETV6-RUNX1+ patients
Approximately 25% of patients diagnosed with paediatric pre-B ALL will present with the t(12;21) ETV6-RUNX1 translocation, which makes it the most frequent genetic insult [106]. The leukaemia blasts from pre-B ALL patients are CD10+, unlike t(4;11)+, and express CD19 and CD34 [107]. The t(12;21) translocation involves the fusion of two transcription factors essential for haematopoiesis: ETV6 and RUNX1. Both genes are expressed in HSCs and more mature haematopoietic progenitors such as LMPP, CLP, GMP and PreMegE [108]. This leads to the formation of a chimeric transcription factor that contains the protein–protein interaction domain of ETV6 (PNT), located on the N-terminal side, with almost the entire RUNX1 gene. The ETV6-RUNX1 chimeric transcription factor does not contain the DNA-binding site of ETV6 (the ETS domain). t(12;21)+ patients have a good prognosis and the risk of relapse is low compared with t(4;11)+ patients. Nearly all ETV6-RUNX1+ pre-B ALL patients can be cured with the minimal residual disease/MRD-guided treatment regimen [109]. Similar to t(4;11) MLL-AF4, the t(12;21) ETV6-RUNX1 originates in utero. Studies in twins found that the genomic breakpoint of the ETV6-RUNX1 chromosomal translocation was identical [110], suggesting that the pre-leukaemic clone reached the other twin via a shared placenta. However, only around 1% of newborns that harbour the t(12;21) will develop leukaemia [111], suggesting that the development of t(12;21) pre-B ALL requires additional mutations. It has been reported that RAG-mediated deletions is one of the main mutational processes [98]. This leads to chromosomal deletions in various genomic loci including ETV6, PAX5, BTG1, LIMD1, NR3C2, ATM, AIM1 and BLIMP1 [112, 113]. A genome-wide association study also identified TP63 (rs17505102) and PTPRJ (rs920590) as germline susceptibility loci in childhood t(12;21) ETV6-RUNX1 pre-B ALL [114]. Around 15% of t(12;21)+ patients harbour an activation mutation in NSD2, which could lead to an increase in H3K36 dimethylation [115]. Further studies also confirmed that t(12;21) ETV6-RUNX1 pre-B ALL is associated with hypermethylation compared to other subtypes such as t(1;19) E2A-PBX1, which has been associated with hypomethylation in the same study [116]. Interestingly, ETV6-RUNX1+ patients also show a preserved global H4 acetylation, which is a favourable prognostic factor [117]. A recent study described a group of patients with an ETV6-RUNX1-like gene expression signature, some of whom also harboured deletions in ETV6 and IKZF1 [118]. This paper additionally identified a new subset of paediatric pre-B ALL patients harbouring a DUX4-rearrangement. This leads to an overexpression of DUX4, a homeobox-containing protein, and a loss of function of the ERG transcription factor [119].
ETV6 and RUNX1: transcription factors vital for embryonic and adult HSCs
In 1997, Wang et al. published the generation of an ETV6 knock-out mouse line [120], with the complete knock-out resulting in embryonic lethality at E10.5 due to an absence of angiogenesis in the yolk sac and apoptosis in specific intra-embryonic cell types. In 2010, Ciau-Uitz et al. showed in Xenopus embryos that Etv6 is essential for the specification of HSCs and the arterial identity of the dorsal aorta through the regulation of Vegfa expression [121]. This study places ETV6 as an essential cell-extrinsic regulator of HSC emergence. In adult blood cells, Runx1 is expressed in transplantable HSCs, myeloid cells and in some lymphoid cells [122], but there is an absolute requirement for the presence of Runx1 only during the initial generation of HSCs during embryogenesis [37]. The lack of Runx1 does not affect yolk sac erythropoiesis, but it severely compromises definitive haematopoiesis [123].
In 1998, Wang et al. investigated the contribution of Etv6 −/− embryonic stem cells in mouse chimeras [124]. They found that Etv6 is dispensable for foetal liver haematopoiesis, but crucially required for adult bone marrow haematopoiesis from the stage of initial bone marrow colonisation. Specifically, they found that Etv6 −/− ES cells do not contribute to adult myelopoiesis or erythropoiesis, and the formation of B cells is also impaired in the absence of Etv6. In 2004, Hock et al. modulated the expression of Etv6 in different blood cell types by crossing mice harbouring a conditional (floxed) Etv6 allele with mice expressing CD19-Cre (B cell), Lck-Cre (T-cell) and Gata1-Cre (erythrocytes and megakaryocytes) [125]. They did not observe changes in the proportions of B and T cells when Etv6 was excised specifically in B or T cells, but found a significant reduction in the frequency of megakaryocytes. Using Mx1-Cre, however, led to a total disruption of adult HSC activity, suggesting that Etv6 is essential for HSC survival, while haematopoietic progenitors further downstream are largely Etv6-independent [125].
In 2015, Rasighaemi et al. inhibited the expression of Etv6 during zebrafish development using two distinct morpholinos [126]. The authors noted changes in primitive and definitive haematopoiesis during embryonic development, notably a decrease in erythrocytes and myeloid cells alongside an increase in lymphopoiesis. Together, those studies highlight the essential role for ETV6 in both the emergence of HSCs during embryonic development (cell-extrinsically) and their maintenance in adult tissues (cell-intrinsically). In addition, they show that Etv6, at least in zebrafish, can affect lymphopoiesis, which is interesting given that the complete inactivation of ETV6 is a frequent genetic event in t(12;21)+ pre-B ALL.
The biology of t(12;21) ETV6-RUNX1 pre-B ALL
The ETV6-RUNX1 fusion gene possesses the PNT domain of ETV6, which is a transcriptional repressor of the Ets family. The PNT domain is the protein interaction domain and recruits proteins like NCOR, SIN3A and histone deacetylases to inhibit transcription [127]. The use of HDAC inhibitors confirmed that the recruitment of histone deacetylases by the PNT domain was part of the leukaemogenesis process, and this treatment led to an accumulation of cells in the G0/G1 phase [128]. Importantly, it affects RUNX1 target genes. For example, ETV6-RUNX1 can inhibit the transcriptional activation of MCSFR, normally promoted by RUNX1 or repressed by ETV6, and this inhibition requires the RUNX1 and C/EBPa consensus sequences [129]. ETV6-RUNX1 was also shown to have a dominant-negative effect on ETV6-mediated transcription, with increasing amounts of ETV6-RUNX1 inhibiting the ETV6-mediated transcriptional repression of stromelysin-1 [130]. Other transcriptional targets of ETV6 include SPHK1, PTGER4 and CLIC5, which can promote survival and proliferation of leukaemia cell lines [131, 132]. Interestingly, ETV6-RUNX1 can also activate the expression of RAC1, which phosphorylates and activates STAT3, leading to a constitutive activation of C-MYC [133]. Accordingly, the inhibition of STAT3 activity through specific inhibitors induced cell death in the pre-leukaemia cell, and shRNA directed against Stat3 led to a longer latency in pre-B ALL development.
Similar to t(4;11) MLL-AF4, many mouse models have been developed to assess the role of ETV6-RUNX1 in disease development. In 2001, Andreasson et al. published a transgenic mouse model, in which the human ETV6-RUNX1 fusion gene was inserted into the locus of the immunoglobulin heavy chain enhancer/promoter EµVHP [134]. This promoter is active in pre-B cells, so the expression of ETV6-RUNX1 was initiated in the population of leukaemia blast cells observed in patients. The mice did not display any signs of haematological malignancy even though the expression of ETV6-RUNX1 was detected in the bone marrow and spleen. This is the first study to suggest that ETV6-RUNX1 alone is not sufficient to drive the development of pre-B ALL.
Bernardin et al. used a transduction approach to study the role of ETV6-RUNX1 in wild-type and Cdkn2a−/− bone marrow cells, followed by transplantation [135]. The alpha form of Cdkn2a (p16INK4a) binds to cyclin-dependent kinase Cdk4 and Cdk6 to inhibit their kinase activity [136]. This leads to a lack of phosphorylation of pRB and a G1 arrest. The beta form of Cdkn2a (p19ARF) activates the p53-MDM2 pathway leading to a G1 cell cycle arrest in both G1 and G2/M [137]. Bernardin et al. found that mice developed an acute leukaemia with a similar penetrance in transduced wild-type and Cdkn2a−/− cells, but the latency was shorter when Cdkn2a was deleted. Li et al. crossed a Tie2-Cre with an inducible ETV6-RUNX1 transgenic mouse, thus targeting expression to endothelial and haematopoietic cells. These mice were then bred to a Cdkn2a-null mouse to assess the contribution of Cdkn2a to leukaemogenesis. Mice that expressed ETV6-RUNX1 and lacked Cdkn2a developed pre-B ALL with a high frequency, and the latency was further reduced with irradiation [138]. Considering that Cdkn2a expression is lost in about 25% of t(12;21)+ pre-B ALL patients, both studies suggest that the inactivation of Cdkn2a is an event that contributes to accelerate leukaemia development [139].
Tsuzuki et al. used a similar approach to Bernardin et al. and transplanted bone marrow cells transduced with a retrovirus expressing ETV6-RUNX1 to investigate the first stages of disease development [140]. They found that ETV6-RUNX1 increased ckit+ haematopoietic stem and progenitor cells, but also increased the proportion of pre-pro-B and pro-B cells in the bone marrow. This B cell differentiation arrest was not complete since there were still pre-B and mature B cells being formed from ETV6-RUNX1+ bone marrow cells. The myeloid and erythroid compartments were largely unaffected. This study suggests that ETV6-RUNX1 can contribute to a differentiation arrest during B lymphoid development, but its presence alone is not sufficient for a complete arrest.
Similarly, Fischer et al. transduced early haematopoietic progenitors with ETV6-RUNX1 and investigated the B cell differentiation arrest and the incidence of leukaemia development in their cohort [141]. Even though their mice did not get sick, the ETV6-RUNX1 transduced cells formed more pre-B cells. When ETV6-RUNX1 expression was higher, the transduced cells started to express ckit and the CD11b myeloid marker. It is plausible that the fine-tuning of ETV6-RUNX1 expression is crucial for restricting the differentiation program towards the B lineage fate. This phenotype has lately been attributed to the transactivation domain of ETV6-RUNX1, which is part of the RUNX1 part of the fusion gene [142].
Similar to t(4;11) MLL-AF4 pro-B ALL, xenotransplantation has been used to assess the role of ETV6-RUNX1 in a human cellular context [143]. The CD34+CD38−CD19+ fraction of t(12;21) ETV6-RUNX1+ patient pre-B ALL bone marrow was transplanted into severe combined immunodeficient mice (NOD/SCID). This population was able to engraft and form B cell colonies that could serially replate. They were also resistant to apoptotic stimuli induced by camptothecin, Fas-L and melphalan.
Schindler et al. used a different approach and inserted the human RUNX1 gene into the mouse Etv6 locus to generate a fusion gene that, like in patients, is under the control of endogenous Etv6 gene regulatory elements [144]. The expression of the Etv6-RUNX1 gene was activated with Gata1-Cre to target early haematopoiesis in the embryo. About two-thirds of the embryos survived an early lethality and showed no obvious phenotype apart from a mild B cell expansion even though foetal liver cells failed to reconstitute the lymphoid compartment upon transplantation. A failure of Etv6-RUNX1-expressing HSCs to differentiate down the lymphoid lineage was further confirmed using Mx1-Cre and Vav-Cre induction.
Kantner et al. created a mouse model of ETV6-RUNX1 by knocking the human ETV6-RUNX1 into the CD19 locus [145]. This led to the expression of ETV6-RUNX1 under the control of the CD19 promoter, which will target committed B lymphoid cells exclusively. The expression level of ETV6-RUNX1 was similar to ETV6, suggesting that it recapitulates the expression level found in leukaemia blasts from patients. The transgenic mice showed similar proportions of the Hardy B cell fractions along the differentiation pathway in the bone marrow and a slight, but not significant, increase of immature B cells in the spleen. Even though the mice did not get sick, pro-B cells showed an elevated cellular level of reactive oxygen species, which translated into an accumulation of DNA damage. This study unravelled a new function of the ETV6-RUNX1 gene as a mediator of DNA damage, a step necessary for disease progression.
Another transduction approach targeted immature (Lineage− or ckit+) cells or pro-B cells from the foetal liver to study the early changes conferred by the ETV6-RUNX1 fusion gene in leukaemogenesis [146]. One retroviral vector conferred high expression of ETV6-RUNX1 while the other vector conferred lower expression, similar to what is observed in patients. The cells with the lower expression of ETV6-RUNX1 could form more and bigger B lymphoid colonies, whereas high expression of ETV6-RUNX1 had a negative impact on B cell development. The ETV6-RUNX1 low-expressing cells also showed an enhanced replating potential, demonstrating that low expression of ETV6-RUNX1 conferred self-renewal to pro-B cells. Interestingly, this phenotype was specific to foetal cells and was not recapitulated in adult cells. Therefore, similar to what is reported for MLL-AF4, it is crucial to maintain an expression level similar to what is observed in patients and to initiate the expression of the fusion gene at the right stage of development.
Several studies investigated the involvement of additional mutations in t(12;21) ETV6-RUNX1 leukaemogenesis. The ASEF gene, a RAC specific guanine nucleotide exchange factor, was reported to be overexpressed in t(12;21) ETV6-RUNX1 pre-B ALL, but not in other haematological malignancies [147]. Its overexpression led to increased B cell potential both in vitro and in vivo, but was not sufficient to induce pre-B ALL (not even in the presence of ETV6-RUNX1). To study t(12;21) in the context of accumulating mutations, van der Weyden et al. generated a mouse model that expressed the Etv6-RUNX1 fusion gene and the Sleeping Beauty transpose under the control of the Etv6 promoter [148]. Around 20% of the mice developed pre-B ALL with a latency of approximately 1 year, which compared to the models mentioned previously and which can be attributed to the presence of additional mutations. By sequencing the genomic DNA from sick mice, they found mutations in genes that correlated with patient data, one of which was Ikzf1 (Ikaros). The deletion of Ikzf1 has been reported in pre-B ALL and leads to an increased expression of HSC genes and reduced expression of B cell genes [149]. It is essential for pre-B cell differentiation [150], so its inactivation would favour the differentiation arrest observed in pre-B ALL. The same group then replaced the Sleeping Beauty transposase component with a Pax5 haploinsufficiency [151]. PAX5 is often deleted or mutated in t(12;21) ETV6-RUNX1 pre-B ALL, with the mutations leading to decreased transcriptional activity [152]. Mice developed a transplantable B cell ALL that was similar to their previous model [148]. Both studies confirmed that t(12;21) ETV6-RUNX1 pre-B ALL requires additional mutations to progress to a full-blown leukaemia. Interestingly, Martìn-Lorenzo et al. also showed that the haploinsufficiency of Pax5 alone can lead to the development of pre-B ALL when the immunological system of the mouse is challenged by an environment that contains pathogens [153]. Both the study by van der Weyden et al. and the one by Martìn-Lorenzo et al. suggested that somatic mutations in Jak3 could also facilitate leukaemia progression [151, 153]. One of the main oncogenic drivers in t(12;21) ETV6-RUNX1 pre-B ALL is RAG-mediated recombination [154]. In 2015, Swaminathan and Klemm et al. showed that genetic lesions in ETV6-RUNX1+ pre-B ALL are usually mediated by AID or RAG1 [155]. Their activity is also increased through chronic inflammation, highlighting the contribution of an overstimulation of the immune system to leukaemogenesis. Another study measured a ten-fold elevation of RAG1 in ETV6-RUNX1+ patients compared to other subtypes of pre-B ALL and found that AID was more expressed in patients lacking a cytogenetic change [156]. All these studies combined suggest that the expression of ETV6-RUNX1 leads to an arrest in the B cell differentiation, but the complete arrest and proliferation of leukaemia blasts requires additional genetic events such as the loss of Pax5 or Cdkn2a activity, which are potentially mediated by an elevated RAG-mediated recombination. The biological features of t(12;21) ETV6-RUNX1 pre-B ALL are summarised in Fig. 2.
t(12;21) ETV6-RUNX1 leukaemia pre-B acute lymphoblastic leukaemia. The different parts of the ETV6-RUNX1 fusion gene are depicted and include the repression domains of ETV6 with almost the entire RUNX1 gene. The ETV6-RUNX1 fusion gene can alter gene expression by targeting RUNX1- and ETV6-target genes such as MCSFR and stromelysin-1. This leukaemia requires cooperating genetic mutations including the inactivation of ETV6, IKAROS, PAX5, CDKN2A and overexpression of ASEF. The prognosis is excellent and almost all patients are cured under the current therapy regimens
The other usual suspect: t(1;19)(q23;p13) E2A-PBX1
Clinical features of E2A-PBX1+ pre-B ALL
The t(1;19) E2A-PBX1 translocation occurs in approximately 25% of paediatric pre-B ALL. It leads to the fusion of PBX1 exons containing the DNA-binding domain and the homeobox domain with the exons of E2A containing the transactivation domain [157]. Alternative splicing leads to the expression of five E2A-PBX1 proteins, where p85E2A-PBX1 and p77E2A-PBX1 are the dominant isoforms [158]. The transforming activity of both proteins is poor in vitro, but can lead to tumour formation in nude mice. Similar to other pre-B ALL, the leukaemia blast population in patients is CD10+CD19+CD34+ [94]. This subtype of leukaemia has been associated with a poor prognosis, but this is mostly for patients that have a balanced t(1;19) translocation [159].
E2A is crucial for the maturation of B lymphoid cells
The E2A locus, also known as TCF3, expresses two proteins: E12 and E47. Once these helix-loop-helix proteins dimerise, they bind E-box sites in promoters and enhancers of genomic DNA to regulate gene expression [160]. E2A is an important regulator of B cell development. The knock-out of E2a in mice results in a high rate of post-natal death [161, 162]. The surviving embryos suffered from growth retardation and an absence of B lymphoid cells, due to reduced levels of Pax5 in the foetal liver. The remaining lineages (T cells, erythrocytes, granulocytes and macrophages) were unaffected by the knock-out of E2a. The expression of E2a peaks during the pro-B to pre-B cell transition once Scl is turned off, and is subsequently downregulated once IgM expression is turned on [163].
E2A activity is negatively regulated by serum growth factors and cell cycle entry, as well as by the ID proteins. The family of ID proteins has been implicated in cancer development and can inhibit the transcriptional activity of Ets transcription factors as well as members of the paired domain transcription factors such as Pax5 (as reviewed in Ref. [164]). The expression of ID2 and ID3 is significantly reduced in pre-B ALL compared to AML, suggesting that their presence is not favourable to pre-B leukaemia blasts [165]. Interestingly, the activity of E2a is also reduced in the bone marrow and spleen of aging mice [166], at a time when HSCs show a trend towards producing fewer lymphoid cells [167]. Overall, the modulation of E2A activity can lead to perturbation in B cell development, thus explaining its involvement in paediatric pre-B ALL.
PBX1, a member of the Hox family of genes
PBX1 is a TALE-class homeodomain transcription factor that interacts with other members of the HOX family of genes to increase their DNA-binding specificities and affinities [168]. PBX1 is a DNA-binding partner of MEIS1 [169], which is also upregulated in t(4;11)+ MLL-AF4 pro-B ALL. The knock-out of Pbx1 in mouse embryonic development leads to profound anaemia and death by E16 because of severe skeletal defects [170]. The absence of Pbx1 also results in abnormalities in the patterning of the great arteries and cardiac outflow tract during embryonic development [171].
In embryonic haematopoiesis, Pbx1 deficiency causes a decrease in myeloid colony-formation and a severe reduction in the engraftment of HSCs from the foetal liver [170]. Pbx1 is highly expressed on the ventral site of the E11.5 AGM, especially in the mesenchyme, but also in haematopoietic progenitors and individual endothelial cells. It continues to be expressed in foetal liver haematopoietic progenitors at later stages and is vital for maintaining definitive haematopoiesis. Pbx1 also plays an essential role in maintaining the quiescence of long-term adult HSCs and in lymphopoiesis [172, 173]. Therefore, the disruption of Pbx1 activity can compromise self-renewal and lymphoid potential of haematopoietic stem and progenitor cells.
The biology of t(1;19) E2A-PBX1 pre-B ALL
The E2A-PBX1 fusion gene is a chimeric transcription factor that lost its ability to interact with MEIS1, but can still interact with HOX genes [174]. Whereas PBX1 transcriptional activity is normally low, E2A-PBX1 can constitutively activate the transcription of PBX1 targets [175]. The stability of the E2A-PBX1 fusion gene can be decreased by HDM2, which promotes ubiquitination and degradation, and increased through acetylation by GCN5 [176]. The interaction between E2A-PBX1 and GCN5 facilitates the expression of target genes such as WNT16 [177], a secreted signalling protein that can activate the expression of Notch ligands (DIc and DId) required for definitive haematopoiesis [178]. Similar to other B cell leukaemias, several mouse models have been developed to understand the role of E2A-PBX1 in leukaemogenesis.
In their 1993 manuscript, Kamps et al. transduced haematopoietic progenitors from the bone marrow with a retrovirus encoding p85E2A-Pbx1, one of the major isoforms, and transplanted these cells to assess leukaemia development [179]. The mice developed leukaemia 11–30 weeks post-transplantation and presented with splenomegaly and an infiltration of the liver and lungs with proliferating blasts. These blasts did not express markers of B or T-cells, but expressed myeloid markers, suggesting that the mice had developed AML, most of which could also grow as granulocytic sarcomas. Using a similar experimental scheme, Sykes et al. found that early pro-T cells immortalised by E2A-PBX1 can generate a myeloid leukaemia (AML) when injected into mice [180]. They also suggested a co-dependence between Scf and E2A-PBX1 for pro-T cell proliferation, which is similar to the co-dependence of SCF and MEIS1 in AML.
In 1993, Uckun et al. presented the results from xenotransplantations of human t(1;19) pre-B ALL leukaemia blasts into SCID mice [181]. The mice developed pre-B ALL within 7–12 weeks after transplantation, with a phenotype recapitulating the human disease. Interestingly, the engraftment was higher when the pre-B ALL blasts came from a patient that relapsed, whereas the pre-B ALL blasts from patients with no relapse did not engraft as well. This study highlights the relationship between the prognosis in patients and the engraftment ability of leukaemia blasts, suggesting the re-acquisition of stem-cell like features in poor prognosis pre-B ALL.
Dedera et al. generated a transgenic mouse model of t(1;19) E2A-PBX1 in which the expression of the fusion gene is under the control of the immunoglobulin heavy chain (IgH) variable region promoter fused to the IgH enhancer (Eµ) [182]. This targets the expression of E2A-PBX1 exclusively to lymphoid cells. Transgenic mice developed T cell leukaemia or lymphoma by 5 months of age. Before the onset of disease, a reduction in the number of thymocytes and bone marrow B lymphoid progenitors was observed. The remaining thymocytes were larger in size compared with cells from control animals, suggesting higher replication and proliferation, but they also exhibited a higher rate of cell death. This suggested that E2A-PBX1 can induce both proliferation and apoptosis simultaneously.
Through the inactivation of CD3 expression, Bijl et al. developed a mouse model which lacked immature and mature T cells [183]. E2A-PBX1/CD3ε−/− mice developed a pre-B cell leukaemia with a latency of around 400 days that was accompanied by an activation of the Hox locus. This feature is also observed in t(4;11) MLL-AF4 pro-B ALL, and thus suggests that HOX activation is involved in another subtype of pre-B ALL.
Using the same mouse model, Hassawi et al. then investigated the role of Hoxa9 in t(1;19) E2A-PBX1 pre-B ALL [183, 184]. The overexpression of Hoxa9 promoted B cell proliferation and led to faster disease progression in E2A-PBX1/CD3ε−/− mice. This was accompanied by reduced levels of Pax5 and Ebf1 gene expression, and an activation of Flt3. The same group also tested the role of HOXB4 in t(1;19) E2A-PBX1 pre-B ALL [185]. First, they performed retroviral transduction of B220+ cells derived from the bone marrow of E2A-PBX1/CD3ε−/− mice and found that the expression of the HOXB4 gene led to an increased production of pro-B cells in vitro, a phenotype linked to proliferation. These cells also exhibited a higher colony-forming potential compared with control cells. Second, they generated a HOXB4/E2A-PBX1/CD3ε−/− mouse line in which HOXB4 and E2A-PBX1 were co-expressed in lymphoid cells. The latency of the disease was not significantly different between E2A-PBX1/CD3ε−/− and HOXB4/E2A-PBX1/CD3ε−/− mice, but the authors detected a higher number of leukaemia-initiating cells upon HOXB4 expression. These studies suggest that HOXA9 and HOXB4 can participate in B cell leukaemogenesis through different mechanisms.
In a recent xenotransplantation study with the 697 leukaemia cell line derived from a t(1;19)+ pre-B ALL patient [186], the aim was to investigate the role of autophagy in leukaemia development through rapamycin treatment which inhibits mTOR activity, since it is known that the activation of autophagy can downregulate E2A-PBX1 expression [187]. All mice developed pre-B ALL with splenomegaly and hepatomegaly, regardless of the rapamycin treatment. However, the mice that received a rapamycin inhibitor after transplantation (or received cells that had been treated with a rapamycin inhibitor before transplantation) had a longer survival period and showed decreased infiltration of leukaemia blasts in the liver. Therefore, the inhibition of the PI3K/AKT/mTOR pathway may assist in eradicating leukaemia blasts, and there is ongoing work aimed at developing an inhibitor that will be useful in the clinic (as reviewed in Ref. [188]).
In a more recent mouse model, Duque-Afonso et al. used a conditional E2A-PBX1 transgenic mouse line in combination with CD19 or Mb1-driven Cre recombinase to target B cells [189]. They also employed Mx1-Cre to induce the expression by polyI:C injection. All mice developed pre-B ALL characterised by a higher white blood cell count, reduced platelet and red blood cell counts in the peripheral blood and splenomegaly. B cell maturation was found to be blocked at the pro-B to early pre-B transition. One of the most recurrent secondary mutations in those mice affected the activation of the Jak/Stat pathway, which may represent another interesting therapeutic avenue. Similar to t(12;21) ETV6-RUNX1 pre-B ALL, Pax5 haploinsufficiency also accelerated leukaemia onset.
Subsequently, the same group reported hyperphosphorylation of Plcγ2 in E2A-PBX1+ mouse leukaemia blasts from the same model [190]. Plcγ2 is a key enzyme in B cell receptor signalling [191]. This hyperphosphorylation was associated with an increase in proliferation and was mediated by Zap70 (zeta chain associated protein kinase), Lck (lymphocyte-specific tyrosine kinase from the SRC family) and Syk (spleen tyrosine kinase), which are downstream targets of the E2A-PBX1 fusion gene. SYK can also upregulate BCL6, a gene essential for the formation of polyclonal B cells and involved in pre-B cell self-renewal [192]. Leukaemia blasts (pre-BCR+ ALL) can thus hijack the pre-BCR signalling pathway to induce differentiation arrest and avoid clonal extinction, and reactivating this pathway may be an attractive therapeutic target (as reviewed in Ref. [193]). Therefore, they also assessed the clonogenic potential of pre-BCR+ and pre-BCR− mouse leukaemia cells in the presence of an LCK inhibitor (A-770041) or SYK inhibitor (P505-15). Encouragingly, both inhibitors led to a significant decrease in colony-forming activity in vitro and increased the survival in vivo, with pre-BCR+ leukaemia cells being more sensitive to A-770041 and P505-15 compared with pre-BCR− leukaemia cells. Pre-BCR+ leukaemia blasts are further characterised by a constitutive activation of the PI3K-AKT signalling pathway and SRC kinase, as well as a downregulation of STAT5 activity [194]. The activation of pre-BCR is more frequent in patients with PBX1 rearrangement and is rarely seen in patients with a MLL rearrangement, t(12;21) ETV6-RUNX1 or t(9;22) BCR-ABL1.
Very recently, Eldfors et al. reported promising results with idelalisib, a phosphatidylinositol 3-kinase delta (p110δ) inhibitor, in a drug-sensitivity assay with leukaemia blasts from a t(1;19)+ relapse patient [195]. The viability of the leukaemia cells was greatly reduced compared with cells from a healthy donor. It also seemed to be more specific to the E2A-PBX1 fusion gene since leukaemia cell lines harbouring other genetic rearrangements were not as sensitive to idelalisib. P110δ is important for maintaining PI3K activity in mature B cells [196], and since the expression of p110δ seems to be more specific to haematopoietic cells, idelalisib may have minimal off-target effects.
Aurora B kinases are overexpressed in many types of paediatric pre-B ALL, but their expression is even higher in t(1;19) E2A-PBX1+ patients [197]. Interestingly, leukaemia cell lines show a significant decrease in proliferation when the expression of Aurora B kinase is downregulated through shRNAs or LNA-oligos. In addition, primary leukaemia cells from patients are more sensitive to the inhibitor barasertib-HQPA when Aurora B kinases are highly expressed. This is due to an increase in histone 3 phosphorylation at Serine 10, which is heavily phosphorylated during metaphase (as reviewed in Ref. [198]), and results in an accumulation of cells in the G2/M phase of the cell cycle, unable to complete cell division.
Rapamycin and its analogues are showing promising results in phase I and phase I/II clinical trials (NCT00874562 and NCT00081874), and a phase III clinical trial is completed for relapsed chronic lymphocytic leukaemia patients and shows that idelalisib improved their overall survival [199] (see also NCT01539512). This drug could also benefit B-ALL patients [195]. The LCK and SYK inhibitors are still in their preclinical phase. Phase I clinical trial for dasatinib (NCT00438854) and barasertib (NCT00926731, NCT01019161) are also completed. The biological features and therapeutic avenues of t(1;19) E2A-PBX1 pre-B ALL are highlighted in Fig. 3.
t(1;19) E2A-PBX1 pre-B acute lymphoblastic leukaemia. E2A-PBX1 can interact with GCN5 to increase its stability through acetylation or with HDM2 to initiate its degradation through ubiquitination. E2A-PBX1 can constitutively activate the expression of PBX1- target genes, which are expressed at low levels under physiological conditions. Many biological pathways contribute to t(1;19)+ pre-B ALL including the activation of Notch ligands, mTOR/PI3K/AKT, JAK/STAT, AuroraB kinase and the hyperphosphorylation of PLCγ2 through specific kinases. Potential therapeutic agents that target these pathways are shown in red
The famous one: t(9;22) BCR-ABL1/Philadelphia chromosome
Clinical features of t(9;22)+ leukaemia
The Philadelphia (Ph) chromosome is found in about 1–3% of paediatric pre-B ALL, and around 25% of adult patients [200]. BCR-ABL1 (or BCR-ABL) is also the main driver of chronic myelogenous leukaemia (CML) with a risk of blast-crisis [201, 202]. This chromosomal translocation leads to the fusion of the 5′ region of the BCR gene with the sequences upstream of the second exon of the ABL1 gene (almost the entire gene coding for this tyrosine kinase), giving rise to the p190 fusion protein found in approximately 85% of Ph+ paediatric ALL and 50–70% of Ph+ adult ALL [203, 204]. In adult CML, the fusion gene contains a larger portion of BCR, resulting in a 210 kDa fusion protein (p210). P210 can also be found in 15% of paediatric ALL and 30–50% of adult ALL [204]. Both p190 and p210 have constitutive kinase activity that can activate many pathways including RAS, RAC, PI3K/AKT/mTOR, NF-κB and JAK/STAT (as reviewed in Ref. [205]) [206]. P230 can be found in rare cases of CML and has a lower intrinsic kinase activity compared to p190 and p210 [204, 207]. Patients with CML are treated with the ABL1 kinase inhibitor imatinib mesylate, which ensures stable remission. Even though patients with pre-B ALL are less sensitive to tyrosine kinase inhibitors [208], the combination with standard chemotherapy has greatly increased the disease-free survival in both adults [209,210,211] and children [212, 213]. For example, the addition of imatinib increased the 4-year disease-free survival from 59% to 75% in paediatric ALL [214] (see also clinical trial NCT00025415). Recently, it was suggested that the ABL1 kinase inhibitor asciminib (ABL001), in combination with nilotinib, could eradicate the development of chronic myelogenous leukaemia in xenografts [215]. Both inhibitors can target the BCR-ABL1 mutant forms that are resistant to imatinib [216]. ABL001 is currently under clinical trial for t(9;22)+ ALL (NCT03106779). Similar to t(12;21) ETV6-RUNX1 pre-B ALL, a genome-wide association identified a GATA3 susceptibility locus (rs3824662) that increases the risk of ALL [217]. This locus is overrepresented in all patients, but even more in Ph+ ALL adolescents and young adults and relapsed patients [217, 218]. The deletion of IKAROS is another frequent genetic event in t(9;22) BCR-ABL1 pre-B ALL [219]. A recent study by Witkowski et al. suggested that Ikaros has a tumour suppressor activity in Ph+ murine B-ALL and its inactivation is crucial for the maintenance of leukaemia [220].
BCR: a breakpoint cluster region protein with kinase activity
BCR has an intrinsic serine/threonine kinase activity that is responsible for phosphorylating BCR serine and threonine residues [221, 222]. The BCR protein can also be phosphorylated by BCR-ABL1 in the part derived from the first exon [154]. The phosphorylation of Tyrosine 177 of BCR by FPS/FES protein tyrosine kinases leads to the binding of Grb2, an activator of Ras [223]. This interaction is important for p190 BCR-ABL1 leukaemogenesis through GAB2-SHP2 signalling [224]. The phosphorylation of Tyrosine 360 is important for the Bcr-mediated transphosphorylation of casein and histone H1, but also for inhibiting the serine/threonine kinase activity [225].
ABL1: a non-receptor tyrosine kinase involved in B cell development
ABL1 is a member of the c-ABL kinase family and can be found in the cytoplasm as well as in the nucleus where it displays DNA-binding activity. The ablation of the C-terminal part of Abl in mice leads to a reduction in the B cell compartment, especially at the pro-B and pre-B stage [226]. The inhibition of c-ABL through a specific antisense oligonucleotide in CD34+ human bone marrow cells leads to an accumulation of cells in the G0/G1 phase of the cell cycle and a decrease in proliferation [227]. It also causes a significant loss in the formation of granulocyte-macrophage colonies. Abl was shown to activate JAK2 phosphorylation in haematopoietic cells [228], a pathway involved in many haematological malignancies. Similar to other genes involved in leukaemia, ABL1 is involved in haematopoiesis and B-lymphoid development.
Biology of t(9;22) BCR-ABL1 pre-B ALL
In 1990, Heisterkamp et al. published a mouse model in which they used the metallothionein-1 promoter, which is active in most tissues, to express BCR-ABL1 [229, 230]. 60% of their transgenic mice developed an acute lymphoblastic leukaemia similar to paediatric pre-B ALL, while 20% developed a myeloid leukaemia which resembled the blast crisis phase of chronic myelogenous leukaemia. There was no thymus involvement for most animals, and the disease progressed rapidly within 10–58 days after birth. Interestingly, expression of BCR-ABL1 under the control of the BCR promoter leads to embryonic lethality [231].
The activation of the RAS pathway is a frequent feature of oncogenic transformation ([232], see also section on MLL-AF4). The human family of RAS comprises three members that are activated in many cancer types (NRAS, KRAS, HRAS) (reviewed in Ref. [233]). These small GTPases can bind and hydrolyse GTP, and their activation requires a series of post-transcriptional modifications to ensure localisation to the plasma membrane. The first of these is the addition of a farnesyl group to a conserved cysteine residue located in the CAAX motif of the C-terminal region. Without farnesylation, RAS remains in the cytoplasm and is inactive [234]. To determine if this was of therapeutic relevance, Reichert et al. investigated the effect of a farnesyl transferase inhibitor SCH66336, also known as lonafarnib, in the p190 BCR-ABL1 mouse model developed by Heisterkamp et al. [229, 235]. Mice that received SCH66336 had a significantly better survival (up to 250 days) than the vehicle control group or the no treatment group (~56 days). After termination of treatment, some mice developed leukaemia, suggesting that this drug cannot kill the leukaemia stem cells. The phase II clinical trial for lonafarnib is completed and could still benefit patients in combination with other drugs (NCT00034684).
The BCR-ABL1 fusion protein can interact with the p85α regulatory subunit of PI3K, an interaction vital for the proliferation of BCR-ABL1-dependent leukaemia cells [236]. The resultant activation of the PI3K/AKT pathway, which also requires the SH2 domain of BCR-ABL1, results in an upregulation of BCL-2 and C-MYC [237]. Accordingly, deletion of both Pi3k subunits (Pik3r1 and Pik3r2) impaired the transformation of murine bone marrow cells with p190 BCR-ABL1 and led to a loss of leukaemogenic potential [238]. An activation of the mTOR pathway independently of the PI3K/AKT pathway was also detected, suggesting that inhibiting both PI3K and mTOR (with the PI-103 inhibitor or similar), combined with the ABL inhibitor imatinib, could be a potential therapeutic avenue for Ph+ pre-B ALL. PI-103 never reached clinical trial because of its high in vivo metabolism [239], but alternative drugs are being tested in phase I clinical trials such as NVP-BEZ235 (NCT01756118).
In 2010, Henry et al. compared the effect of BCR-ABL1 on the B lymphoid potential of young and aged bone marrow cells [240]. Untransduced bone marrow cells showed a similar myeloid output, but the B lymphoid potential decreased with age. When transduced with p190 BCR-ABL1, cells of both ages displayed a higher engraftment and an expansion of early B cell progenitors, pro-B cells, immature and mature B cells when old bone marrow cells were used as competitor cells. Furthermore, p190 BCR-ABL1-expressing cells showed a higher level of phosphorylated Akt, Stat5 and Erk. The use of old competitor cells in their transplantation experiments, along with young or old bone marrow cells transduced with p190 BCR-ABL1, led to the development of leukaemia. Their study suggested that young competitor cells may not be ideal when modelling B cell leukaemia using transplantation.
In 2011, Notta et al. described a Ph+ ALL xenograft system that allowed the identification of multiple leukaemia-initiating clones in patients [241]. Recipients that were reconstituted with the predominant clone at diagnosis displayed a deletion of CDKN2A/B and fast disease. Interestingly, the xenograft clones constituted a population of leukaemia-initiating cells that did not always correspond to the sample at diagnosis. These results confirmed the heterogeneity of the leukaemia-initiating cell compartment and proposed that the engraftment is mostly due to more aggressive clones. This was also confirmed in another xenograft model by Ebinger et al. in 2016 [69].
Waldron et al. assessed the role of c-Myb and Bmi1 on the development of leukaemia in the p190 BCR-ABL1 mouse model [242]. They found that the loss of one c-Myb allele is sufficient to slow p190 BCR-ABL1 leukaemogenesis and that this is partly due to a decrease in Bmi1 expression. This study showed that activation of the c-Myb–Bmi1 axis is important for maintaining a high level of B-lymphoid clonogenic potential and ensuring disease progression of BCR-ABL1 pre-B ALL.
B cell ALL displays a differentiation arrest at the pro-B to pre-B stage and is associated with constitutive RAG activity, terminal deoxy-transferase expression and ongoing Ig heavy (IgH) chain rearrangement [243, 244]. In addition, the NF-κB transcription factor is constitutively active in ALL cells [245]. Ochodnicka-Mackovicova et al. investigated the relationship between the NF-κB and RAG pathways and noticed that the inhibition of NF-κB leads to an upregulation of RAG expression and increased RAG-dependent DNA damage in a transformed mouse pre-B cell model [246]. In patients, a low expression of RAG1, RAG2 and TDT correlates with higher expression of NF-κB. Since the inhibition of NF-κB can lead to increased RAG-dependent DNA damage in patient leukaemia cells, this therapeutic avenue should be considered with extreme care. Rag-mediated recombination can lead to the deletion of Ikaros in BCR-ABL1+ pre-B ALL, which facilitates leukaemogenesis [219].
The IL7 receptor (IL7R) is crucially involved in lymphoid development and its loss leads to reduced lymphoid cellularity in the peripheral blood [247]. Shochat et al. recently reported gain-of-function mutations in IL7R in paediatric B- and T-ALL through a serine-to-cysteine substitution (Ser185) in the extracellular region, or in-frame insertions and deletions in the transmembrane domain [248]. This resulted in the constitutive activation of the receptor and cytokine-independent growth of progenitor lymphoid cells. IL7R, once activated, can interact with JAK1 and JAK3 to phosphorylate and activate STAT5. The JAK/STAT pathway is often activated in leukaemia blasts of Ph+ pre-B ALL [249, 250]. P190 BCR-ABL1 can mediate the tyrosine phosphorylation of STAT members, and JAK1 and JAK2 have been found to be constitutively activated by mutations in the kinase and pseudokinase domains, which explains the sensitivity of Ph+ leukaemia cells to JAK2 inhibitors [251, 252]. The biological features and therapeutic avenues of t(9;22) BCR-ABL1 pre-B ALL are presented in Fig. 4.
t(9;22) BCR-ABL1 pre-B acute lymphoblastic leukaemia. This leukaemia depends on the hijack of signalling pathways, including the constitutive activation of ABL kinase and the activation of PI3K/AKT/mTOR pathways. These can be targeted by inhibitors of ABL (imatinib), mutant forms of BCR-ABL1 (asciminib/ABL001 and nilotinib) and PI3K/mTOR (PI-103 and NVP-BEZ235). The activation of RAS, IL7R, C-MYB/BMI-1 and the phosphorylation of JAK2 can also contribute to disease. RAS can be inhibited by lonafarnib, a farnesyl transferase inhibitor, and ruxolitinib can inhibit JAK2 activity
BCR-ABL1-like
In 2009, Den Boer et al. constructed a classifier based on gene expression to shed light on the 25% of pre-B ALL cases that are genetically unclassified [253]. This led to the identification of a BCR-ABL1-like phenotype in 15–50% of paediatric cases that harbour deletions in IKZF1, PAX5, EBF1, TCF3 and VPREB1. The BCR-ABL1-like group was then confirmed in 30–40% of adult B-ALL cases [254]. In both age groups, this subtype is associated with a poor response to treatment through resistance to L-asparaginase and daunorubicin, and also a high risk of relapse [253]. In 2014, Roberts et al. published a genome profiling study aimed at identifying genetic alterations that could explain the poor prognosis [255]. They found a number of rearrangements of genes involved in tyrosine kinase signalling (ABL1, ABL2, CRFL2, CSF1R, EPOR, JAK2, NTRK3, PDGFRB, PTK2B, TSLP and TYK2) and mutations in FLT3, IL7R and SH2B3. Some of these rearrangements led to a constitutive activation of STAT5 and a sensitivity to dasatinib (ABL1, ABL2, CSF1R and PDGFRB fusion genes), ruxolitinib (EPOR and JAK2 rearrangements) and crizotinib (ETV6-NTRK3) [255, 256]. Furthermore, the use of tyrosine kinase inhibitors in BCR-ABL1-like patients and refractory cases has shown promising therapeutic results [257,258,259]. The phase II clinical trial for dasatinib is completed (NCT00438854) while the recruitment has started for ruxolitinib (NCT02723994). Ex vivo analysis with primary patient cells are underway for crizotinib (NCT02551718).
Hyperdiploidy
Around 20–30% of paediatric and around 3% of adult B cell leukaemia are associated with a gain in chromosome numbers. There is evidence for a prenatal origin in some patients, but the frequency is low compared to other subtypes such as t(4;11) MLL-AF4 or t(12;21) ETV6-RUNX1 [260]. Patients respond very well to therapeutic drugs or DNA topoisomerase inhibitors due to an overstimulation of the apoptotic response by hyperdiploid leukaemia cells [261, 262]. A significant proportion of patients harbours mutations in FLT3 (~25%), NRAS/KRAS (~15%) or PTNP11 genes (~10%), which are also involved in other paediatric pro- and pre-B ALL [263,264,265]. These mutations are mutually exclusive in most cases [266]. Other frequent abnormalities involve deletions in loci of ETV6, CKDN2A, PAX5 and PAN3 [ 267 ].
Hypodiploidy
Hypodiploidy is a rare subset of B cell leukaemia in both children and adults (1–2%) and is associated with a very poor prognosis. The aetiology is not known, but the gene dosage or unmasking of recessive alleles could be part of the answer (as reviewed in Ref. [268]). Holmfeldt et al. recently described the genomic landscape of hypodiploid ALL using whole-genome and exome sequencing [269]. Around 71% of near-haploid patients (24–31 chromosomes) had mutations in genes leading to the activation of receptor tyrosine kinases and Ras signalling, and 13% presented deletions and one frameshift mutation in IKZF3, a lymphoid transcription factor of the IKAROS family. They also found mutations in TP53 (~90%), IKZF2 (~36%) and RB1 (~41%) in low-hypodiploid patients (32–39 chromosomes). All patients showed activation of Ras- and PI3K-signalling and were sensitive to PI3K inhibitors, highlighting a potential therapeutic avenue. A phase I clinical trial is underway for the dual PI3K/mTOR inhibitor NVP-BEX235 (NCT01756118), and the phase I trial for the PI3K inhibitor GDC-0941 is complete (NCT00876122).
Trisomy 4 and 10
Trisomy of chromosomes 4 and 10 accounts for 20–25% of paediatric pre-B ALL, but is very rare in adults (Table 1). It is associated with an extremely favourable 4-year event-free survival (~96%) compared to around 70% for patients with none or just one trisomy (~70%) [270].
Intrachromosomal amplification of chromosome 21
The involvement of chromosome 21 in leukaemia is present in 2% of cases of paediatric and 11% of adult B-ALL cases (as reviewed in Ref. [271]). Furthermore, children with Down syndrome have a 10–12 fold higher chance of developing an acute leukaemia compared to children without Down syndrome (as reviewed in Ref. [272]). This can lead to an amplification of the Runx1 locus in a subset of patients [273]. It is associated with a poor outcome and requires more intensive chemotherapy (HR COG ALL protocols) [274].
Conclusion
In this review, we aimed at providing an updated report of the field of B cell leukaemia with a particular emphasis on infant/paediatric ALL. In both paediatric and adult patients, the transformation process is usually initiated by a chromosomal translocation, creating a fusion gene with aberrant activity. Some fusion genes will interfere at the transcriptional level (MLL-AF4, ETV6-RUNX1, E2A-PBX1) whereas others will activate signalling pathways that promote oncogenesis (BCR-ABL1). The first stages of transformation will prime the cells to become pre-malignant (pre-leukaemia cells) by inducing a differentiation arrest during B cell development and the acquisition of stem cell features (e.g. upregulation of Hox genes). What we have learned from the numerous mouse models of pro/pre-B ALL is that additional events are necessary to boost the transformation process and bring those pre-leukaemia cells to induce a full-blown leukaemia. These events include genetic mutations that inactivate key players of B cell development (e.g. Pax5, Ikaros, Ebf1) or inhibitors of apoptosis (e.g. BCL-2 and MCL-1). There may also be an activation of genes involved in cell division and proliferation (e.g. CDK6, phosphorylation of PLCγ2). We are only starting to identify and understand the impact of co-drivers on pro/pre-B ALL, and these co-drivers can lead to the development of novel therapeutic avenues. For future studies, it will also be important to include external factors that can recapitulate the human environment. For example, an overstimulation of the immune response can lead to B cell leukaemia in Pax5 haploinsufficient mice, whereas a clean environment does not lead to disease. Similar findings could be true for other factors (e.g. pesticides). It is also essential to understand the role of these mechanisms specifically in leukaemia stem cells as relapse from these therapy-resistant cells is a major clinical problem.
Abbreviations
- AGM:
-
Aorta-gonads-mesonephros
- AML:
-
Acute myeloid leukaemia
- B-ALL:
-
B cell acute lymphoblastic leukaemia
- CLP:
-
Common lymphoid progenitor
- CML:
-
Chronic myelogenous leukaemia
- CMP:
-
Common myeloid progenitor
- ES:
-
Embryonic stem
- GMP:
-
Granulocyte–macrophage progenitor
- HSC:
-
Haematopoietic stem cell
- LMPP:
-
Lymphoid-primed multipotent progenitor
- MPP:
-
Multipotent progenitor
- polyI:C:
-
Polyinosinic/polycytidylic acid
- PreMegE:
-
Premegakaryocytic/erythroid
- VEC:
-
VE-cadherin
References
Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66(1):7–30. doi:10.3322/caac.21332
Perera FP (1996) Molecular epidemiology: insights into cancer susceptibility, risk assessment, and prevention. J Natl Cancer Inst 88(8):496–509
Martin B, Beverely K, Adrianne H (2007) Risk factors for acute leukemia in children: a review. Environ Health Perspect 115(1):138–145. doi:10.1289/ehp.9023
Yeoh E-J, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou X, Li J, Liu H, Pui C-H, Evans WE, Naeve C, Wong L, Downing JR (2002) Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1(2):133–143. doi:10.1016/S1535-6108(02)00032-6
Ma X, Buffler PA, Gunier RB, Dahl G, Smith MT, Reinier K, Reynolds P (2002) Critical windows of exposure to household pesticides and risk of childhood leukemia. Environ Health Perspect 110(9):955–960
Ahlbom A, Day N, Feychting M, Roman E, Skinner J, Dockerty J, Linet M, McBride M, Michaelis J, Olsen JH, Tynes T, Verkasalo PK (2000) A pooled analysis of magnetic fields and childhood leukaemia. Br J Cancer 83(5):692–698. doi:10.1054/bjoc.2000.1376
Roman E, Simpson J, Ansell P, Kinsey S, Mitchell CD, McKinney PA, Birch JM, Greaves M, Eden T (2007) Childhood acute lymphoblastic leukemia and infections in the first year of life: a report from the United Kingdom Childhood Cancer Study. Am J Epidemiol 165(5):496–504. doi:10.1093/aje/kwk039
Kinlen L (1988) Evidence for an infective cause of childhood leukaemia: comparison of a scottish new town with nuclear reprocessing sites in Britain. The Lancet 332(8624):1323–1327. doi:10.1016/S0140-6736(88)90867-7
Greaves M (1988) Speculations on the cause of childhood acute lymphoblastic leukemia. Leukemia 2(2):120–125
Society AC (2014) Cancer facts and figures 2014. American Cancer Society, Atlanta
Hunger SP, Mullighan CG (2015) Acute lymphoblastic leukemia in children. N Engl J Med 373(16):1541–1552. doi:10.1056/NEJMra1400972
Pui C-H, Evans WE (2006) Treatment of acute lymphoblastic leukemia. N Engl J Med 354(2):166–178. doi:10.1056/NEJMra052603
Gale KB, Ford AM, Repp R, Borkhardt A, Keller C, Eden OB, Greaves MF (1997) Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc Natl Acad Sci USA 94(25):13950–13954
Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, Downing JR (2008) Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 322(5906):1377. doi:10.1126/science.1164266
Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP, Tang Z, Zumbo P, Li S, Zavadil J, Levine RL, Cardozo T, Hunger SP, Raetz EA, Evans WE, Morrison DJ, Mason CE, Carroll WL (2013) Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet 45(3):290–294. doi:10.1038/ng.2558
Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, Buetow KH, Pui C-H, Baker SD, Brindle PK, Downing JR (2011) CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 471(7337):235–239. doi:10.1038/nature09727
Thirman MJ, Gill HJ, Burnett RC, Mbangkollo D, McCabe NR, Kobayashi H, Ziemin-van der Poel S, Kaneko Y, Morgan R, Sandberg AA, Chaganti RSK, Larson RA, Le Beau MM, Diaz MO, Rowley JD (1993) Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N Engl J Med 329(13):909–914. doi:10.1056/NEJM199309233291302
Behm FG, Raimondi SC, Frestedt JL, Liu Q, Crist WM, Downing JR, Rivera GK, Kersey JH, Pui CH (1996) Rearrangement of the MLL gene confers a poor prognosis in childhood acute lymphoblastic leukemia, regardless of presenting age. Blood 87(7):2870–2877
Jansen MWJC, Corral L, van der Velden VHJ, Panzer-Grumayer R, Schrappe M, Schrauder A, Marschalek R, Meyer C, den Boer ML, Hop WJC, Valsecchi MG, Basso G, Biondi A, Pieters R, van Dongen JJM (2007) Immunobiological diversity in infant acute lymphoblastic leukemia is related to the occurrence and type of MLL gene rearrangement. Leukemia 21(4):633–641. doi:10.1038/sj.leu.2404578
Pui CH, Frankel LS, Carroll AJ, Raimondi SC, Shuster JJ, Head DR, Crist WM, Land VJ, Pullen DJ, Steuber CP (1991) Clinical characteristics and treatment outcome of childhood acute lymphoblastic leukemia with the t(4;11)(q21;q23): a collaborative study of 40 cases. Blood 77(3):440–447
Schwartz S, Rieder H, Schlager B, Burmeister T, Fischer L, Thiel E (2003) Expression of the human homologue of rat NG2 in adult acute lymphoblastic leukemia: close association with MLL rearrangement and a CD10(−)/CD24(−)/CD65s(+)/CD15(+) B-cell phenotype. Leukemia 17(8):1589–1595. doi:10.1038/sj.leu.2402989
Burmeister T, Meyer C, Schwartz S, Hofmann J, Molkentin M, Kowarz E, Schneider B, Raff T, Reinhardt R, Gökbuget N, Hoelzer D, Thiel E, Marschalek R (2009) The MLL recombinome of adult CD10-negative B-cell precursor acute lymphoblastic leukemia: results from the GMALL study group. Blood 113(17):4011–4015. doi:10.1182/blood-2008-10-183483
Bueno C, Montes R, Martin L, Prat I, Hernandez MC, Orfao A, Menendez P (2008) NG2 antigen is expressed in CD34+ HPCs and plasmacytoid dendritic cell precursors: is NG2 expression in leukemia dependent on the target cell where leukemogenesis is triggered? Leukemia 22(8):1475–1478. doi:10.1038/leu.2008.134
Hotfilder M, Röttgers S, Rosemann A, Schrauder A, Schrappe M, Pieters R, Jürgens H, Harbott J, Vormoor J (2005) Leukemic stem cells in childhood high-risk ALL/t(9;22) and t(4;11) are present in primitive lymphoid-restricted CD34+ CD19− cells. Cancer Res 65(4):1442–1449. doi:10.1158/0008-5472.CAN-04-1356
Menendez P, Catalina P, Rodríguez R, Melen GJ, Bueno C, Arriero M, García-Sánchez F, Lassaletta A, García-Sanz R, García-Castro J (2009) Bone marrow mesenchymal stem cells from infants with MLL-AF4+ acute leukemia harbor and express the MLL-AF4 fusion gene. J Exp Med 206(13):3131–3141. doi:10.1084/jem.20091050
Bueno C, Montes R, Melen GJ, Ramos-Mejia V, Real PJ, Ayllon V, Sanchez L, Ligero G, Gutierrez-Aranda I, Fernandez AF, Fraga MF, Moreno-Gimeno I, Burks D, del Carmen Plaza-Calonge M, Rodriguez-Manzaneque JC, Menendez P (2012) A human ESC model for MLL-AF4 leukemic fusion gene reveals an impaired early hematopoietic-endothelial specification. Cell Res 22(6):986–1002. doi:10.1038/cr.2012.4
Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL (2007) Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res 67(15):7275. doi:10.1158/0008-5472.CAN-06-2369
Godfrey L, Kerry J, Thorne R, Repapi E, Davies JOJ, Tapia M, Ballabio E, Hughes JR, Geng H, Konopleva M, Milne TA (2017) MLL-AF4 binds directly to a BCL-2 specific enhancer and modulates H3K27 acetylation. Exp Hematol 47:64–75. doi:10.1016/j.exphem.2016.11.003
Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, Showalter HD, Murai MJ, Belcher AM, Hartley T, Hess JL, Cierpicki T (2012) Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol 8(3):277–284. doi:10.1038/nchembio.773
He S, Malik B, Borkin D, Miao H, Shukla S, Kempinska K, Purohit T, Wang J, Chen L, Parkin B, Malek SN, Danet-Desnoyers G, Muntean AG, Cierpicki T, Grembecka J (2016) Menin-MLL inhibitors block oncogenic transformation by MLL-fusion proteins in a fusion partner-independent manner. Leukemia 30(2):508–513. doi:10.1038/leu.2015.144
Senisterra G, Wu H, Allali-Hassani A, Wasney Gregory A, Barsyte-Lovejoy D, Dombrovski L, Dong A, Nguyen Kong T, Smil D, Bolshan Y, Hajian T, He H, Seitova A, Chau I, Li F, Poda G, Couture J-F, Brown Peter J, Al-Awar R, Schapira M, Arrowsmith Cheryl H, Vedadi M (2013) Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem J 449(Pt 1):151–159. doi:10.1042/BJ20121280
Cao F, Townsend Elizabeth C, Karatas H, Xu J, Li L, Lee S, Liu L, Chen Y, Ouillette P, Zhu J, Hess Jay L, Atadja P, Lei M, Qin Zhaohui S, Malek S, Wang S, Dou Y (2014) Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol Cell 53(2):247–261. doi:10.1016/j.molcel.2013.12.001
Čermáková K, Tesina P, Demeulemeester J, El Ashkar S, Méreau H, Schwaller J, Řezáčová P, Veverka V, De Rijck J (2014) Validation and structural characterization of the LEDGF/p75–MLL interface as a new target for the treatment of MLL-dependent leukemia. Cancer Res 74(18):5139–5151. doi:10.1158/0008-5472.CAN-13-3602
Yu BD, Hess JL, Horning SE, Brown GAJ, Korsmeyer SJ (1995) Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378(6556):505–508
Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ (1998) MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci 95(18):10632–10636. doi:10.1038/378505a0
Medvinsky A, Dzierzak E (1996) Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86(6):897–906. doi:10.1016/S0092-8674(00)80165-8
North TE, de Bruijn MFTR, Stacy T, Talebian L, Lind E, Robin C, Binder M, Dzierzak E, Speck NA (2002) Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 16(5):661–672. doi:10.1016/S1074-7613(02)00296-0
Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ (1997) Defects in yolk sac hematopoiesis in Mll-null embryos. Blood 90(5):1799–1806
Palis J, Robertson S, Kennedy M, Wall C, Keller G (1999) Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126(22):5073–5084
Yagi H, Deguchi K, Aono A, Tani Y, Kishimoto T, Komori T (1998) Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood 92(1):108–117
Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ (2004) Definitive hematopoiesis requires the Mixed-Lineage Leukemia gene. Dev Cell 6(3):437–443. doi:10.1016/S1534-5807(04)00061-9
McMahon KA, Hiew SYL, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJM (2007) Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 1(3):338–345. doi:10.1016/j.stem.2007.07.002
Melko M, Douguet D, Bensaid M, Zongaro S, Verheggen C, Gecz J, Bardoni B (2011) Functional characterization of the AFF (AF4/FMR2) family of RNA-binding proteins: insights into the molecular pathology of FRAXE intellectual disability. Hum Mol Genet 20(10):1873–1885. doi:10.1093/hmg/ddr069
Chao S-H, Price DH (2001) Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 276(34):31793–31799. doi:10.1074/jbc.M102306200
Chen CS, Hilden JM, Frestedt J, Domer PH, Moore R, Korsmeyer SJ, Kersey JH (1993) The chromosome 4q21 gene (AF-4/FEL) is widely expressed in normal tissues and shows breakpoint diversity in t(4;11)(q21;q23) acute leukemia. Blood 82(4):1080–1085
Frestedt JL, Hilden JM, Moore RO, Kersey JH (1996) Differential expression of AF4/FEL mRNA in human tissues. Genet Anal Biomol Eng 12(3):147–149. doi:10.1016/1050-3862(95)00127-1
Isnard P, Coré N, Naquet P, Djabali M (2000) Altered lymphoid development in mice deficient for the mAF4 proto-oncogene. Blood 96(2):705–710
Mak AB, Nixon AML, Moffat J (2012) The mixed lineage leukemia (MLL) fusion-associated gene AF4 promotes CD133 transcription. Cancer Res 72(8):1929–1934
Okuda H, Kanai A, Ito S, Matsui H, Yokoyama A (2015) AF4 uses the SL1 components of RNAP1 machinery to initiate MLL fusion- and AEP-dependent transcription. Nat Commun 6:8869. doi:10.1038/ncomms9869
Okuda H, Takahashi S, Takaori-Kondo A, Yokoyama A (2016) TBP loading by AF4 through SL1 is the major rate-limiting step in MLL fusion-dependent transcription. Cell Cycle 15(20):2712–2722. doi:10.1080/15384101.2016.1222337
Scholz B, Kowarz E, Rössler T, Ahmad K, Steinhilber D, Marschalek R (2015) AF4 and AF4N protein complexes: recruitment of P-TEFb kinase, their interactome and potential functions. Am J Blood Res 5(1):10–24
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale A-L, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinsk M, Jager N, Jones DTW, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt ANJ, Valdes-Mas R, van Buuren MM, van’t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Australian Pancreatic Cancer Genome Initiative, Consortium IBC, Consortium IM-S, PedBrain ICGC, Zucman-Rossi J, Andrew Futreal P, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR (2013) Signatures of mutational processes in human cancer. Nature 500 (7463):415–421. doi:10.1038/nature12477
Meshorer E, Misteli T (2006) Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol 7(7):540–546. doi:10.1038/nrm1938
Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, Nakitandwe J, Holmfeldt L, Parker M, Easton J, Huether R, Kriwacki R, Rusch M, Wu G, Li Y, Mulder H, Raimondi S, Pounds S, Kang G, Shi L, Becksfort J, Gupta P, Payne-Turner D, Vadodaria B, Boggs K, Yergeau D, Manne J, Song G, Edmonson M, Nagahawatte P, Wei L, Cheng C, Pei D, Sutton R, Venn NC, Chetcuti A, Rush A, Catchpoole D, Heldrup J, Fioretos T, Lu C, Ding L, Pui C-H, Shurtleff S, Mullighan CG, Mardis ER, Wilson RK, Gruber TA, Zhang J, Downing JR, for The St. Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome Project (2015) The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet 47(4):330–337. doi:10.1038/ng.3230
Trentin L, Bresolin S, Giarin E, Bardini M, Serafin V, Accordi B, Fais F, Tenca C, De Lorenzo P, Valsecchi MG, Cazzaniga G, Gt Kronnie, Basso G (2016) Deciphering KRAS and NRAS mutated clone dynamics in MLL-AF4 paediatric leukaemia by ultra deep sequencing analysis. Sci Rep 6:34449. doi:10.1038/srep34449
Prieto C, Stam RW, Agraz-Doblas A, Ballerini P, Camos M, Castaño J, Marschalek R, Bursen A, Varela I, Bueno C, Menendez P (2016) Activated KRAS cooperates with MLL-AF4 to promote extramedullary engraftment and migration of cord Blood CD34+ HSPC but is insufficient to initiate leukemia. Cancer Res 76(8):2478–2489. doi:10.1158/0008-5472.CAN-15-2769
Tamai H, Miyake K, Takatori M, Miyake N, Yamaguchi H, Dan K, Shimada T, Inokuchi K (2011) Activated K-Ras protein accelerates human MLL/AF4-induced leukemo-lymphomogenicity in a transgenic mouse model. Leukemia 25(5):888–891. doi:10.1038/leu.2011.15
Chen W, Li Q, Hudson WA, Kumar A, Kirchhof N, Kersey JH (2006) A murine Mll-AF4 knock-in model results in lymphoid and myeloid deregulation and hematologic malignancy. Blood 108(2):669–677. doi:10.1182/blood-2005-08-3498
Metzler M, Forster A, Pannell R, Arends MJ, Daser A, Lobato MN, Rabbitts TH (2006) A conditional model of MLL-AF4 B-cell tumourigenesis using invertor technology. Oncogene 25(22):3093–3103. doi:10.1038/sj.onc.1209636
Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, Kutok JL, Kung AL, Armstrong SA (2008) H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 14(5):355–368. doi:10.1016/j.ccr.2008.10.001
Ito R, Takahashi T, Katano I, Ito M (2012) Current advances in humanized mouse models. Cell Mol Immunol 9(3):208–214. doi:10.1038/cmi.2012.2
Montes R, Ayllón V, Gutierrez-Aranda I, Prat I, Hernández-Lamas MC, Ponce L, Bresolin S, te Kronnie G, Greaves M, Bueno C, Menendez P (2011) Enforced expression of MLL-AF4 fusion in cord blood CD34+ cells enhances the hematopoietic repopulating cell function and clonogenic potential but is not sufficient to initiate leukemia. Blood 117(18):4746–4758. doi:10.1182/blood-2010-12-322230
Lin S, Luo Roger T, Ptasinska A, Kerry J, Assi Salam A, Wunderlich M, Imamura T, Kaberlein Joseph J, Rayes A, Althoff Mark J, Anastasi J, O’Brien Maureen M, Meetei Amom R, Milne Thomas A, Bonifer C, Mulloy James C, Thirman Michael J (2016) Instructive role of MLL-fusion proteins revealed by a model of t(4;11) pro-B acute lymphoblastic leukemia. Cancer Cell 30(5):737–749. doi:10.1016/j.ccell.2016.10.008
Babovic S, Eaves CJ (2014) Hierarchical organization of fetal and adult hematopoietic stem cells. Exp Cell Res 329(2):185–191. doi:10.1016/j.yexcr.2014.08.005
Barrett Neil A, Malouf C, Kapeni C, Bacon Wendi A, Giotopoulos G, Jacobsen Sten Eirik W, Huntly Brian J, Ottersbach K (2016) Mll-AF4 confers enhanced self-renewal and lymphoid potential during a restricted window in development. Cell Reports 16(4):1039–1054. doi:10.1016/j.celrep.2016.06.046
Frame JM, McGrath KE, Palis J (2013) Erythro-myeloid progenitors: “Definitive” hematopoiesis in the conceptus prior to the emergence of hematopoietic stem cells. Blood Cells Mol Dis 51(4):220–225. doi:10.1016/j.bcmd.2013.09.006
Böiers C, Carrelha J, Lutteropp M, Luc S, Green Joanna CA, Azzoni E, Woll Petter S, Mead Adam J, Hultquist A, Swiers G, Perdiguero Elisa G, Macaulay Iain C, Melchiori L, Luis Tiago C, Kharazi S, Bouriez-Jones T, Deng Q, Pontén A, Atkinson D, Jensen Christina T, Sitnicka E, Geissmann F, Godin I, Sandberg R, de Bruijn Marella FTR, Jacobsen Sten Eirik W (2013) Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell 13(5):535–548. doi:10.1016/j.stem.2013.08.012
Greaves MF (1993) Stem cell origins of leukaemia and curability. Br J Cancer 67(3):413–423
Ebinger S, Özdemir EZ, Ziegenhain C, Tiedt S, Castro Alves C, Grunert M, Dworzak M, Lutz C, Turati VA, Enver T, Horny H-P, Sotlar K, Parekh S, Spiekermann K, Hiddemann W, Schepers A, Polzer B, Kirsch S, Hoffmann M, Knapp B, Hasenauer J, Pfeifer H, Panzer-Grümayer R, Enard W, Gires O, Jeremias I (2016) Characterization of rare, dormant, and therapy-resistant cells in acute lymphoblastic leukemia. Cancer Cell 30(6):849–862. doi:10.1016/j.ccell.2016.11.002
Cox CV, Evely RS, Oakhill A, Pamphilon DH, Goulden NJ, Blair A (2004) Characterization of acute lymphoblastic leukemia progenitor cells. Blood 104(9):2919–2925. doi:10.1182/blood-2004-03-0901
le Viseur C, Hotfilder M, Bomken S, Wilson K, Röttgers S, Schrauder A, Rosemann A, Irving J, Stam RW, Shultz LD, Harbott J, Jürgens H, Schrappe M, Pieters R, Vormoor J (2008) In childhood acute lymphoblastic leukemia, blasts at different stages of immunophenotypic maturation have stem cell properties. Cancer Cell 14(1):47–58. doi:10.1016/j.ccr.2008.05.015
Aoki Y, Watanabe T, Saito Y, Kuroki Y, Hijikata A, Takagi M, Tomizawa D, Eguchi M, Eguchi-Ishimae M, Kaneko A, Ono R, Sato K, Suzuki N, Fujiki S, Koh K, Ishii E, Shultz LD, Ohara O, Mizutani S, Ishikawa F (2015) Identification of CD34+ and CD34− leukemia-initiating cells in MLL-rearranged human acute lymphoblastic leukemia. Blood 125(6):967–980. doi:10.1182/blood-2014-03-563304
Bardini M, Woll PS, Corral L, Luc S, Wittmann L, Ma Z, Lo Nigro L, Basso G, Biondi A, Cazzaniga G, Jacobsen SEW (2015) Clonal variegation and dynamic competition of leukemia-initiating cells in infant acute lymphoblastic leukemia with MLL rearrangement. Leukemia 29(1):38–50. doi:10.1038/leu.2014.154
Peham M, Panzer S, Fasching K, Haas OA, Fisher S, Marschalek R, Gadner H, Panzer-Grümayer RE (2002) Low frequency of clonotypic Ig and T-cell receptor gene rearrangements in t(4;11) infant acute lymphoblastic leukaemia and its implication for the detection of minimal residual disease. Br J Haematol 117(2):315–321. doi:10.1046/j.1365-2141.2002.03428.x
Caslini C, Shilatifard A, Yang L, Hess JL (2000) The amino terminus of the mixed lineage leukemia protein (MLL) promotes cell cycle arrest and monocytic differentiation. Proc Natl Acad Sci USA 97(6):2797–2802. doi:10.1073/pnas.040574897
Thomas M, Geßner A, Vornlocher H-P, Hadwiger P, Greil J, Heidenreich O (2005) Targeting MLL-AF4 with short interfering RNAs inhibits clonogenicity and engraftment of t(4;11)-positive human leukemic cells. Blood 106(10):3559–3566. doi:10.1182/blood-2005-03-1283
Lin S, Luo RT, Shrestha M, Thirman MJ, Mulloy JC (2017) The full transforming capacity of MLL-Af4 is interlinked with lymphoid lineage commitment. Blood. doi:10.1182/blood-2017-04-777185
Dou L, Zheng D, Li J, Li Y, Gao L, Wang L, Yu L (2012) Methylation-mediated repression of microRNA-143 enhances MLL-AF4 oncogene expression. Oncogene 31(4):507–517. doi:10.1038/onc.2011.248
Chillon MC, Gomez-Casares MT, Lopez-Jorge CE, Rodriguez-Medina C, Molines A, Sarasquete ME, Alcoceba M, Miguel JDGS, Bueno C, Montes R, Ramos F, Rodriguez JN, Giraldo P, Ramirez M, Garcia-Delgado R, Fuster JL, Gonzalez-Diaz M, Menendez P (2012) Prognostic significance of FLT3 mutational status and expression levels in MLL-AF4+ and MLL-germline acute lymphoblastic leukemia. Leukemia 26(11):2360–2366. doi:10.1038/leu.2012.161
Wang GG, Pasillas MP, Kamps MP (2005) Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood 106(1):254–264. doi:10.1182/blood-2004-12-4664
Montes R, Ayllon V, Prieto C, Bursen A, Prelle C, Romero-Moya D, Real PJ, Navarro-Montero O, Chillon C, Marschalek R, Bueno C, Menendez P (2014) Ligand-independent FLT3 activation does not cooperate with MLL-AF4 to immortalize/transform cord blood CD34+ cells. Leukemia 28(3):666–674. doi:10.1038/leu.2013.346
Kowarz E, Burmeister T, Lo Nigro L, Jansen MWJC, Delabesse E, Klingebiel T, Dingermann T, Meyer C, Marschalek R (2007) Complex MLL rearrangements in t(4;11) leukemia patients with absent AF4.MLL fusion allele. Leukemia 21(6):1232–1238. doi:10.1038/sj.leu.2404686
Benedikt A, Baltruschat S, Scholz B, Bursen A, Arrey TN, Meyer B, Varagnolo L, Muller AM, Karas M, Dingermann T, Marschalek R (2011) The leukemogenic AF4-MLL fusion protein causes P-TEFb kinase activation and altered epigenetic signatures. Leukemia 25(1):135–144. doi:10.1038/leu.2010.249
Castaño J, Herrero AB, Bursen A, González F, Marschalek R, Gutiérrez NC, Menendez P (2016) Expression of MLL-AF4 or AF4-MLL fusions does not impact the efficiency of DNA damage repair. Oncotarget 7(21):30440–30452. doi:10.18632/oncotarget.8938
Ahmad K, Scholz B, Capelo R, Schweighöfer I, Kahnt AS, Marschalek R, Steinhilber D (2015) AF4 and AF4-MLL mediate transcriptional elongation of 5-lipoxygenase mRNA by 1, 25-dihydroxyvitamin D(3). Oncotarget 6(28):25784–25800. doi:10.18632/oncotarget.4703
Wang D, DuBois RN (2010) Eicosanoids and cancer. Nat Rev Cancer 10(3):181–193. doi:10.1038/nrc2809
Runarsson G, Liu A, Mahshid Y, Feltenmark S, Pettersson A, Klein E, Björkholm M, Claesson H-E (2005) Leukotriene B4 plays a pivotal role in CD40-dependent activation of chronic B lymphocytic leukemia cells. Blood 105(3):1274–1279. doi:10.1182/blood-2004-07-2546
Bursen A, Schwabe K, Rüster B, Henschler R, Ruthardt M, Dingermann T, Marschalek R (2010) The AF4·MLL fusion protein is capable of inducing ALL in mice without requirement of MLL·AF4. Blood 115(17):3570–3579. doi:10.1182/blood-2009-06-229542
Liang D-C, Shih L-Y, Fu J-F, Li H-Y, Wang H-I, Hung I-J, Yang C-P, Jaing T-H, Chen S-H, Liu H-C (2006) K-ras mutations and N-ras mutations in childhood acute leukemias with or without mixed-lineage leukemia gene rearrangements. Cancer 106(4):950–956. doi:10.1002/cncr.21687
Ng M-HJ, Ng RK, Kong C-T, Jin D-Y, Chan L-C (2010) Activation of Ras-dependent Elk-1 activity by MLL-AF4 family fusion oncoproteins. Exp Hematol 38(6):481–488. doi:10.1016/j.exphem.2010.03.014
Bernt Kathrin M, Zhu N, Sinha Amit U, Vempati S, Faber J, Krivtsov Andrei V, Feng Z, Punt N, Daigle A, Bullinger L, Pollock Roy M, Richon Victoria M, Kung Andrew L, Armstrong Scott A (2011) MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20(1):66–78. doi:10.1016/j.ccr.2011.06.010
Daigle SR, Iwanowics D, Johnston LD, Therkelsen CA, Smith JJ, Moyer MP, Copeland RA, Olhava EJ, Scott MP, Pollock RM, Raimondi A (2013) DOT1L Inhibitor EPZ-5676 displays synergistic antiproliferative activity in combination with standard of care drugs or DNA hypomethylating agents in MLL-rearranged leukemia cells. Blood 122(21):3930
Gutierrez MI, Siraj AK, Bhargava M, Ozbek U, Banavali S, Chaudhary MA, El Solh H, Bhatia K (2003) Concurrent methylation of multiple genes in childhood ALL: correlation with phenotype and molecular subgroup. Leukemia 17(9):1845–1850. doi:10.1038/sj.leu.2403060
Lawrence HJ, Christensen J, Fong S, Hu Y-L, Weissman I, Sauvageau G, Humphries RK, Largman C (2005) Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood 106(12):3988–3994. doi:10.1182/blood-2005-05-2003
Azcoitia V, Aracil M, Martínez-A C, Torres M (2005) The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev Biol 280(2):307–320. doi:10.1016/j.ydbio.2005.01.004
Copley MR, Babovic S, Benz C, Knapp DJHF, Beer PA, Kent DG, Wohrer S, Treloar DQ, Day C, Rowe K, Mader H, Kuchenbauer F, Humphries RK, Eaves CJ (2013) The Lin28b–let-7–Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol 15(8):916–925. doi:10.1038/ncb2783
Wilkinson Adam C, Ballabio E, Geng H, North P, Tapia M, Kerry J, Biswas D, Roeder Robert G, Allis CD, Melnick A, de Bruijn Marella FTR, Milne Thomas A (2013) RUNX1 is a key target in t(4;11) leukemias that contributes to gene activation through an AF4-MLL complex interaction. Cell Rep 3(1):116–127. doi:10.1016/j.celrep.2012.12.016
Wu Z, Eguchi-Ishimae M, Yagi C, Iwabuki H, Gao W, Tauchi H, Inukai T, Sugita K, Ishii E, Eguchi M (2015) HMGA2 as a potential molecular target in KMT2A-AFF1-positive infant acute lymphoblastic leukaemia. Br J Haematol 171(5):818–829. doi:10.1111/bjh.13763
Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, Croce CM, Nakamura T, Canaani E, Young RA (2008) Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev 22(24):3403–3408. doi:10.1101/gad.1741408
Benito Juliana M, Godfrey L, Kojima K, Hogdal L, Wunderlich M, Geng H, Marzo I, Harutyunyan Karine G, Golfman L, North P, Kerry J, Ballabio E, Chonghaile Triona N, Gonzalo O, Qiu Y, Jeremias I, Debose L, O’Brien E, Ma H, Zhou P, Jacamo R, Park E, Coombes Kevin R, Zhang N, Thomas Deborah A, O’Brien S, Kantarjian Hagop M, Leverson Joel D, Kornblau Steven M, Andreeff M, Müschen M, Zweidler-McKay Patrick A, Mulloy James C, Letai A, Milne Thomas A, Konopleva M (2015) MLL-rearranged acute lymphoblastic leukemias activate BCL-2 through H3K79 methylation and are sensitive to the BCL-2-specific antagonist ABT-199. Cell Rep 13(12):2715–2727. doi:10.1016/j.celrep.2015.12.003
Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan W-I, Robson SC, C-w Chung, Hopf C, Savitski MM, Huthmacher C, Gudgin E, Lugo D, Beinke S, Chapman TD, Roberts EJ, Soden PE, Auger KR, Mirguet O, Doehner K, Delwel R, Burnett AK, Jeffrey P, Drewes G, Lee K, Huntly BJP, Kouzarides T (2011) Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478(7370):529–533. doi:10.1038/nature10509
Laurenti E, Frelin C, Xie S, Ferrari R, Dunant Cyrille F, Zandi S, Neumann A, Plumb I, Doulatov S, Chen J, April C, Fan J-B, Iscove N, Dick John E (2015) CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell 16(3):302–313. doi:10.1016/j.stem.2015.01.017
Chaidos A, Caputo V, Karadimitris A (2015) Inhibition of bromodomain and extra-terminal proteins (BET) as a potential therapeutic approach in haematological malignancies: emerging preclinical and clinical evidence. Ther Adv Hematol 6(3):128–141. doi:10.1177/2040620715576662
Morera L, Lübbert M, Jung M (2016) Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenetics 8(1):57. doi:10.1186/s13148-016-0223-4
Cierpicki T, Grembecka J (2014) Challenges and opportunities in targeting the menin-MLL interaction. Future Med Chem 6(4):447–462. doi:10.4155/fmc.13.214
Romana SP, Poirel H, Leconiat M, Flexor MA, Mauchauffe M, Jonveaux P, Macintyre EA, Berger R, Bernard OA (1995) High frequency of t(12;21) in childhood B-lineage acute lymphoblastic leukemia. Blood 86(11):4263–4269
Chiaretti S, Zini G, Bassan R (2014) Diagnosis and subclassification of acute lymphoblastic leukemia. Mediterr J Hematol Infect Dis 6(1):e2014073. doi:10.4084/MJHID.2014.073
Moignard V, Macaulay IC, Swiers G, Buettner F, Schütte J, Calero-Nieto FJ, Kinston S, Joshi A, Hannah R, Theis FJ, Jacobsen SE, de Bruijn MF, Göttgens B (2013) Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat Cell Biol 15(4):363–372. doi:10.1038/ncb2709
Bhojwani D, Pei D, Sandlund JT, Jeha S, Ribeiro RC, Rubnitz JE, Raimondi SC, Shurtleff S, Onciu M, Cheng C, Coustan-Smith E, Bowman WP, Howard SC, Metzger ML, Inaba H, Leung W, Evans WE, Campana D, Relling MV, Pui CH (2012) ETV6-RUNX1-positive childhood acute lymphoblastic leukemia: improved outcome with contemporary therapy. Leukemia 26(2):265–270. doi:10.1038/leu.2011.227
Ford AM, Bennett CA, Price CM, Bruin MCA, Van Wering ER, Greaves M (1998) Fetal origins of the TEL-AML1 fusion gene in identical twins with leukemia. Proc Natl Acad Sci 95(8):4584–4588
Lausten-Thomsen U, Madsen HO, Vestergaard TR, Hjalgrim H, Nersting J, Schmiegelow K (2011) Prevalence of t(12;21)[ETV6-RUNX1]-positive cells in healthy neonates. Blood 117(1):186–189. doi:10.1182/blood-2010-05-282764
Montpetit A, Boily G, Sinnett D (2002) A detailed transcriptional map of the chromosome 12p12 tumour suppressor locus. Eur J Hum Genet 10(1):62–71. doi:10.1038/sj.ejhg.5200766
Tsuzuki S, Karnan S, Horibe K, Matsumoto K, Kato K, Inukai T, Goi K, Sugita K, Nakazawa S, Kasugai Y, Ueda R, Seto M (2007) Genetic abnormalities involved in t(12;21) TEL–AML1 acute lymphoblastic leukemia: analysis by means of array-based comparative genomic hybridization. Cancer Sci 98(5):698–706. doi:10.1111/j.1349-7006.2007.00443.x
Ellinghaus E, Stanulla M, Richter G, Ellinghaus D, te Kronnie G, Cario G, Cazzaniga G, Horstmann M, Panzer Grümayer R, Cavé H, Trka J, Cinek O, Teigler-Schlegel A, ElSharawy A, Häsler R, Nebel A, Meissner B, Bartram T, Lescai F, Franceschi C, Giordan M, Nürnberg P, Heinzow B, Zimmermann M, Schreiber S, Schrappe M, Franke A (2012) Identification of germline susceptibility loci in ETV6-RUNX1-rearranged childhood acute lymphoblastic leukemia. Leukemia 26(5):902–909. doi:10.1038/leu.2011.302
Jaffe JD, Wang Y, Chan HM, Zhang J, Huether R, Kryukov GV, H-eC Bhang, Taylor JE, Hu M, Englund NP, Yan F, Wang Z, Robert McDonald Iii E, Wei L, Ma J, Easton J, Yu Z, deBeaumount R, Gibaja V, Venkatesan K, Schlegel R, Sellers WR, Keen N, Liu J, Caponigro G, Barretina J, Cooke VG, Mullighan C, Carr SA, Downing JR, Garraway LA, Stegmeier F (2013) Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet 45(11):1386–1391. doi:10.1038/ng.2777
Gabriel AS, Lafta FM, Schwalbe EC, Nakjang S, Cockell SJ, Iliasova A, Enshaei A, Schwab C, Rand V, Clifford SC, Kinsey SE, Mitchell CD, Vora A, Harrison CJ, Moorman AV, Strathdee G (2015) Epigenetic landscape correlates with genetic subtype but does not predict outcome in childhood acute lymphoblastic leukemia. Epigenetics 10(8):717–726. doi:10.1080/15592294.2015.1061174
Janczar K, Janczar S, Pastorczak A, Mycko K, Paige AJW, Zalewska-Szewczyk B, Wagrowska-Danilewicz M, Danilewicz M, Mlynarski W (2015) Preserved global histone H4 acetylation linked to ETV6-RUNX1 fusion and PAX5 deletions is associated with favorable outcome in pediatric B-cell progenitor acute lymphoblastic leukemia. Leuk Res 39(12):1455–1461. doi:10.1016/j.leukres.2015
Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, Olsson L, Orsmark-Pietras C, von Palffy S, Askmyr M, Rissler M, Schrappe M, Cario G, Castor A, Pronk CJH, Behrendtz M, Mitelman F, Johansson B, Paulsson K, Andersson AK, Fontes M, Fioretos T (2016) Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun 7:11790. doi:10.1038/ncomms11790
Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, Wu G, Li Y, Wei L, Iacobucci I, Liu Y, Qu C, Wen J, Edmonson M, Payne-Turner D, Kaufmann KB, S-i Takayanagi, Wienholds E, Waanders E, Ntziachristos P, Bakogianni S, Wang J, Aifantis I, Roberts KG, Ma J, Song G, Easton J, Mulder HL, Chen X, Newman S, Ma X, Rusch M, Gupta P, Boggs K, Vadodaria B, Dalton J, Liu Y, Valentine ML, Ding L, Lu C, Fulton RS, Fulton L, Tabib Y, Ochoa K, Devidas M, Pei D, Cheng C, Yang J, Evans WE, Relling MV, Pui C-H, Jeha S, Harvey RC, Chen IML, Willman CL, Marcucci G, Bloomfield CD, Kohlschmidt J, Mrozek K, Paietta E, Tallman MS, Stock W, Foster MC, Racevskis J, Rowe JM, Luger S, Kornblau SM, Shurtleff SA, Raimondi SC, Mardis ER, Wilson RK, Dick JE, Hunger SP, Loh ML, Downing JR, Mullighan CG, the St. Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome Project (2016) Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet 48(12):1481–1489. doi:10.1038/ng.3691
Wang LC, Kuo F, Fujiwara Y, Gilliland DG, Golub TR, Orkin SH (1997) Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J 16(14):4374–4383
Ciau-Uitz A, Pinheiro P, Gupta R, Enver T, Patient R (2010) Tel1/ETV6 specifies blood stem cells through the agency of VEGF signaling. Dev Cell 18(4):569–578. doi:10.1016/j.devcel.2010.02.009
North TE, Stacy T, Matheny CJ, Speck NA, de Bruijn MFTR (2004) Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells 22(2):158–168. doi:10.1634/stemcells.22-2-158
Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84(2):321–330. doi:10.1016/S0092-8674(00)80986-1
Wang LC, Swat W, Fujiwara Y, Davidson L, Visvader J, Kuo F, Alt FW, Gilliland DG, Golub TR, Orkin SH (1998) The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev 12(15):2392–2402
Hock H, Meade E, Medeiros S, Schindler JW, Valk PJ, Fujiwara Y, Orkin SH (2004) Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev 18(19):2336–2341. doi:10.1101/gad.1239604
Rasighaemi P, Onnebo SMN, Liongue C, Ward AC (2015) ETV6 (TEL1) regulates embryonic hematopoiesis in zebrafish. Haematologica 100(1):23–31. doi:10.3324/haematol.2014.104091
Hollenhorst PC, McIntosh LP, Graves BJ (2011) Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem 80:437–471. doi:10.1146/annurev.biochem.79.081507.103945
Starkova J, Madzo J, Cario G, Kalina T, Ford A, Zaliova M, Hrusak O, Trka J (2007) The identification of (ETV6)/RUNX1-regulated genes in lymphopoiesis using histone deacetylase inhibitors in ETV6/RUNX1-positive lymphoid leukemic cells. Clin Cancer Res 13(6):1726–1735. doi:10.1158/1078-0432.CCR-06-2569
Fears S, Gavin M, Zhang DE, Hetherington C, Ben-David Y, Rowley JD, Nucifora G (1997) Functional characterization of ETV6 and ETV6/CBFA2 in the regulation of the MCSFR proximal promoter. Proc Natl Acad Sci USA 94(5):1949–1954
Gunji H, Waga K, Nakamura F, Maki K, Sasaki K, Nakamura Y, Mitani K (2004) TEL/AML1 shows dominant-negative effects over TEL as well as AML1. Biochem Biophys Res Commun 322(2):623–630. doi:10.1016/j.bbrc.2004.07.169
Malouf C, Lagace K, Drullion C, Langlois S, Larose J, Kritikou EA, Sirard C, Drouin S, Sinnett D (2016) Novel transcriptional targets of ETV6, a transcription factor frequently altered in childhood pre-B acute lymphoblastic leukemia. Cancer Sci Open Res Access 3(1):1–11
Neveu B, Spinella J-F, Richer C, Lagacé K, Cassart P, Lajoie M, Jananji S, Drouin S, Healy J, Hickson GRX, Sinnett D (2016) CLIC5: a novel ETV6 target gene in childhood acute lymphoblastic leukemia. Haematologica 101(12):1534–1543. doi:10.3324/haematol.2016.149740
Mangolini M, de Boer J, Walf-Vorderwülbecke V, Pieters R, den Boer ML, Williams O (2013) STAT3 mediates oncogenic addiction to TEL-AML1 in t(12;21) acute lymphoblastic leukemia. Blood 122(4):542–549. doi:10.1182/blood-2012-11-465252
Andreasson P, Schwaller J, Anastasiadou E, Aster J, Gilliland DG (2001) The expression of ETV6/CBFA2 (TEL/AML1) is not sufficient for the transformation of hematopoietic cell lines in vitro or the induction of hematologic disease in vivo. Cancer Genet Cytogenet 130(2):93–104. doi:10.1016/S0165-4608(01)00518-0
Bernardin F, Yang Y, Cleaves R, Zahurak M, Cheng L, Civin CI, Friedman AD (2002) TEL-AML1, expressed from t(12;21) in human acute lymphocytic leukemia, induces acute leukemia in mice. Cancer Res 62(14):3904–3908
Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81(3):323–330. doi:10.1016/0092-8674(95)90385-2
Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, Peters G (1998) The alternative product from the human CDKN2A locus, p14 ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J 17(17):5001–5014. doi:10.1093/emboj/17.17.5001
Li M, Jones L, Gaillard C, Binnewies M, Ochoa R, Garcia E, Lam V, Wei G, Yang W, Lobe C, Hermiston M, Passegue E, Kogan SC (2013) Initially disadvantaged, TEL-AML1 cells expand and initiate leukemia in response to irradiation and cooperating mutations. Leukemia 27(7):1570–1573. doi:10.1038/leu.2013.15
Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui C-H, Relling MV, Evans WE, Shurtleff SA, Downing JR (2007) Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446(7137):758–764. doi:10.1038/nature05690
Tsuzuki S, Seto M, Greaves M, Enver T (2004) Modeling first-hit functions of the t(12;21) TEL-AML1 translocation in mice. Proc Natl Acad Sci USA 101(22):8443–8448. doi:10.1073/pnas.0402063101
Fischer M, Schwieger M, Horn S, Niebuhr B, Ford A, Roscher S, Bergholz U, Greaves M, Lohler J, Stocking C (2005) Defining the oncogenic function of the TEL//AML1 (ETV6//RUNX1) fusion protein in a mouse model. Oncogene 24(51):7579–7591. doi:10.1038/sj.onc.1208931
Morrow M, Samanta A, Kioussis D, Brady HJM, Williams O (2007) TEL-AML1 preleukemic activity requires the DNA binding domain of AML1 and the dimerization and corepressor binding domains of TEL. Oncogene 26(30):4404–4414. doi:10.1038/sj.onc.1210227
Hong D, Gupta R, Ancliff P, Atzberger A, Brown J, Soneji S, Green J, Colman S, Piacibello W, Buckle V, Tsuzuki S, Greaves M, Enver T (2008) Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science 319(5861):336–339. doi:10.1126/science.1150648
Schindler JW, Van Buren D, Foudi A, Krejci O, Qin J, Orkin SH, Hock H (2009) TEL-AML1 corrupts hematopoietic stem cells to persist in the bone marrow and initiate leukemia. Cell Stem Cell 5(1):43–53. doi:10.1016/j.stem.2009.04.019
Kantner H-P, Warsch W, Delogu A, Bauer E, Esterbauer H, Casanova E, Sexl V, Stoiber D (2013) ETV6/RUNX1 induces reactive oxygen species and drives the accumulation of DNA damage in B cells. Neoplasia 15(11):1292–1300. doi:10.1593/neo.131310
Tsuzuki S, Seto M (2013) TEL (ETV6)-AML1 (RUNX1) initiates self-renewing fetal pro-B cells in association with a transcriptional program shared with embryonic stem cells in mice. Stem Cells 31(2):236–247. doi:10.1002/stem.1277
Lyons R, Williams O, Morrow M, Sebire N, Hubank M, Anderson J (2010) The RAC specific guanine nucleotide exchange factor Asef functions downstream from TEL-AML1 to promote leukaemic transformation. Leuk Res 34(1):109–115. doi:10.1016/j.leukres.2009.06.032
van der Weyden L, Giotopoulos G, Rust AG, Matheson LS, van Delft FW, Kong J, Corcoran AE, Greaves MF, Mullighan CG, Huntly BJ, Adams DJ (2011) Modeling the evolution of ETV6-RUNX1-induced B-cell precursor acute lymphoblastic leukemia in mice. Blood 118(4):1041–1051
Mullighan CG, Su X, Zhang J, Radtke I, Phillips LAA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, Harvey RC, Chen I-M, Clifford RJ, Carroll WL, Reaman G, Bowman WP, Devidas M, Gerhard DS, Yang W, Relling MV, Shurtleff SA, Campana D, Borowitz MJ, Pui C-H, Smith M, Hunger SP, Willman CL, Downing JR, Group tCsO (2009) Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 360(5):470–480. doi:10.1056/NEJMoa0808253
Heizmann B, Kastner P, Chan S (2013) Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J Exp Med 210(13):2823–2832. doi:10.1084/jem.20131735
van der Weyden L, Giotopoulos G, Wong K, Rust AG, Robles-Espinoza CD, Osaki H, Huntly BJ, Adams DJ (2015) Somatic drivers of B-ALL in a model of ETV6-RUNX1; Pax5+/− leukemia. BMC Cancer 15:585. doi:10.1186/s12885-015-1586-1
Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, Wechsler J, Yang J, Hayes J, Klein RJ, Zhang J, Wei L, Wu G, Rusch M, Nagahawatte P, Ma J, Chen S-C, Song G, Cheng J, Meyers P, Bhojwani D, Jhanwar S, Maslak P, Fleisher M, Littman J, Offit L, Rau-Murthy R, Fleischut MH, Corines M, Murali R, Gao X, Manschreck C, Kitzing T, Murty VV, Raimondi SC, Kuiper RP, Simons A, Schiffman JD, Onel K, Plon SE, Wheeler DA, Ritter D, Ziegler DS, Tucker K, Sutton R, Chenevix-Trench G, Li J, Huntsman DG, Hansford S, Senz J, Walsh T, Lee M, Hahn CN, Roberts KG, King M-C, Lo SM, Levine RL, Viale A, Socci ND, Nathanson KL, Scott HS, Daly M, Lipkin SM, Lowe SW, Downing JR, Altshuler D, Sandlund JT, Horwitz MS, Mullighan CG, Offit K (2013) A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet 45(10):1226–1231. doi:10.1038/ng.2754
Martin-Lorenzo A, Hauer J, Vicente-Duenas C, Auer F, Gonzalez-Herrero I, Garcia-Ramirez I, Ginzel S, Thiele R, Constantinescu SN, Bartenhagen C, Dugas M, Gombert M, Schafer D, Blanco O, Mayado A, Orfao A, Alonso-Lopez D, De Las Rivas J, Cobaleda C, Garcia-Cenador MB, Garcia-Criado FJ, Sanchez-Garcia I, Borkhardt A (2015) Infection exposure is a causal factor in B-precursor acute lymphoblastic leukemia as a result of Pax5 inherited susceptibility. Cancer Discov 5(12):1328–1343. doi:10.1158/2159-8290.CD-15-0892
Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J, Alexandrov LB, Van Loo P, Cooke SL, Marshall J, Martincorena I, Hinton J, Gundem G, van Delft FW, Nik-Zainal S, Jones DR, Ramakrishna M, Titley I, Stebbings L, Leroy C, Menzies A, Gamble J, Robinson B, Mudie L, Raine K, O’Meara S, Teague JW, Butler AP, Cazzaniga G, Biondi A, Zuna J, Kempski H, Muschen M, Ford AM, Stratton MR, Greaves M, Campbell PJ (2014) RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet 46(2):116–125. doi:10.1038/ng.2874
Swaminathan S, Klemm L, Park E, Papaemmanuil E, Ford A, Kweon S-M, Trageser D, Hasselfeld B, Henke N, Mooster J, Geng H, Schwarz K, Kogan SC, Casellas R, Schatz DG, Lieber MR, Greaves MF, Muschen M (2015) Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol 16(7):766–774. doi:10.1038/ni.3160
Heinäniemi M, Vuorenmaa T, Teppo S, Kaikkonen MU, Bouvy-Liivrand M, Mehtonen J, Niskanen H, Zachariadis V, Laukkanen S, Liuksiala T, Teittinen K, Lohi O (2016) Transcription-coupled genetic instability marks acute lymphoblastic leukemia structural variation hotspots. eLife 5:e13087. doi:10.7554/eLife.13087
Nourse J, Mellentin JD, Galili N, Wilkinson J, Stanbridge E, Smith SD, Cleary ML (1990) Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell 60(4):535–545. doi:10.1016/0092-8674(90)90657-Z
Kamps MP, Look T, Baltimore D (1991) The human t(1;19) translocation in pre-B ALL produces multiple nuclear E2A-Pbx1 fusion proteins with differing transforming potentials. Genes Dev 5(3):358–368
Uckun FM, Sensel MG, Sather HN, Gaynon PS, Arthur DC, Lange BJ, Steinherz PG, Kraft P, Hutchinson R, Nachman JB, Reaman GH, Heerema NA (1998) Clinical significance of translocation t(1;19) in childhood acute lymphoblastic leukemia in the context of contemporary therapies: a report from the Children’s Cancer Group. J Clin Oncol 16(2):527–535. doi:10.1200/jco.1998.16.2.527
Aronheim A, Shiran R, Rosen A, Walker MD (1993) The E2A gene product contains two separable and functionally distinct transcription activation domains. Proc Natl Acad Sci 90(17):8063–8067
Zhuang Y, Soriano P, Weintraub H (1994) The helix–loop–helix gene E2A is required for B cell formation. Cell 79(5):875–884. doi:10.1016/0092-8674(94)90076-0
Bain G, Maandag ECR, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, van der Valk M, te Riele HPJ, Berns A, Murre C (1994) E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79(5):885–892. doi:10.1016/0092-8674(94)90077-9
Herblot S, Aplan PD, Hoang T (2002) Gradient of E2A activity in B-cell development. Mol Cell Biol 22(3):886–900. doi:10.1128/MCB.22.3.886-900.2002
Lasorella A, Benezra R, Iavarone A (2014) The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer 14(2):77–91. doi:10.1038/nrc3638
May AM, Frey A-V, Bogatyreva L, Benkisser-Petersen M, Hauschke D, Lübbert M, Wäsch R, Werner M, Hasskarl J, Lassmann S (2014) ID2 and ID3 protein expression mirrors granulopoietic maturation and discriminates between acute leukemia subtypes. Histochem Cell Biol 141(4):431–440. doi:10.1007/s00418-013-1169-7
Frasca D, Nguyen D, Riley RL, Blomberg BB (2003) Decreased E12 and/or E47 transcription factor activity in the bone marrow as well as in the spleen of aged mice. J Immunol 170(2):719–726. doi:10.4049/jimmunol.170.2.719
Benz C, Copley Michael R, Kent David G, Wohrer S, Cortes A, Aghaeepour N, Ma E, Mader H, Rowe K, Day C, Treloar D, Brinkman Ryan R, Eaves Connie J (2017) Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell 10(3):273–283. doi:10.1016/j.stem.2012.02.007
Chang CP, Brocchieri L, Shen WF, Largman C, Cleary ML (1996) Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol Cell Biol 16(4):1734–1745
Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB (2001) Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development 128(21):4139–4151
DiMartino JF, Selleri L, Traver D, Firpo MT, Rhee J, Warnke R, Gorman S, Weissman IL, Cleary ML (2001) The Hox cofactor and proto-oncogene Pbx1 is required for maintenance of definitive hematopoiesis in the fetal liver. Blood 98(3):618–626. doi:10.1182/blood.V98.3.618
Chang C-P, Stankunas K, Shang C, Kao S-C, Twu KY, Cleary ML (2008) Pbx1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development 135(21):3577–3586. doi:10.1242/dev.022350
Ficara F, Murphy MJ, Lin M, Cleary ML (2008) Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell 2(5):484–496. doi:10.1016/j.stem.2008.03.004
Ficara F, Crisafulli L, Lin C, Iwasaki M, Smith KS, Zammataro L, Cleary ML (2013) Pbx1 restrains myeloid maturation while preserving lymphoid potential in hematopoietic progenitors. J Cell Sci 126(14):3181–3191. doi:10.1242/jcs.125435
Chang CP, Jacobs Y, Nakamura T, Jenkins NA, Copeland NG, Cleary ML (1997) Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol Cell Biol 17(10):5679–5687
Lu Q, Wright DD, Kamps MK (1994) Fusion with E2A converts the Pbx1 homeodomain protein into a constitutive transcriptional activator in human leukemias carrying the t(1;19) translocation. Mol Cell Biol 14(6):3938–3948. doi:10.1128/MCB.14.6.3938
Holmlund T, Lindberg MJ, Grander D, Wallberg AE (2013) GCN5 acetylates and regulates the stability of the oncoprotein E2A-PBX1 in acute lymphoblastic leukemia. Leukemia 27(3):578–585. doi:10.1038/leu.2012.265
McWhirter JR, Neuteboom STC, Wancewicz EV, Monia BP, Downing JR, Murre C (1999) Oncogenic homeodomain transcription factor E2A-Pbx1 activates a novel WNT gene in pre-B acute lymphoblastoid leukemia. Proc Natl Acad Sci 96(20):11464–11469. doi:10.1073/pnas.96.20.11464
Clements WK, Kim AD, Ong KG, Moore JC, Lawson ND, Traver D (2011) A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature 474(7350):220–224. doi:10.1038/nature10107
Kamps MP, Baltimore D (1993) E2A-Pbx1, the t(1;19) translocation protein of human pre-B-cell acute lymphocytic leukemia, causes acute myeloid leukemia in mice. Mol Cell Biol 13(1):351–357
Sykes DB, Kamps MP (2004) E2a/Pbx1 induces the rapid proliferation of stem cell factor-dependent murine pro-T cells that cause acute T-lymphoid or myeloid leukemias in mice. Mol Cell Biol 24(3):1256–1269. doi:10.1128/MCB.24.3.1256-1269.2004
Uckun FM, Downing JR, Gunther R, Chelstrom LM, Finnegan D, Land VJ, Borowitz MJ, Carroll AJ, Crist WM (1993) Human t(1;19)(q23;p13) pre-B acute lymphoblastic leukemia in mice with severe combined immunodeficiency. Blood 81(11):3052–3062
Dedera DA, Waller EK, LeBrun DP, Sen-Majumdar A, Stevens ME, Barsh GS, Cleary ML (1993) Chimeric homeobox gene E2A-PBX1 induces proliferation, apoptosis, and malignant lymphomas in transgenic mice. Cell 74(5):833–843. doi:10.1016/0092-8674(93)90463-Z
Bijl J, Sauvageau M, Thompson A, Sauvageau G (2005) High incidence of proviral integrations in the Hoxa locus in a new model of E2a–PBX1-induced B-cell leukemia. Genes Dev 19(2):224–233. doi:10.1101/gad.1268505
Hassawi M, Shestakova EA, Fournier M, C-ét Leber-Gali, Vaisson G, Hélïs Frison, Sinnett D, Vidal R, Thompson A, Bijl JJ (2014) Hoxa9 collaborates with E2A-PBX1 in mouse B cell leukemia in association with Flt3 activation and decrease of B cell gene expression. Dev Dyn 243(1):145–158. doi:10.1002/dvdy.24056
Fournier M, Savoie-Rondeau I, Larochelle F, Hassawi M, Shestakova EA, Roy DC, Bijl JJ (2014) Inability of HOXB4 to enhance self-renewal of malignant B cells: favorable profile for the expansion of autologous hematopoietic stem cells. Exp Hematol 42(7):526–535. doi:10.1016/j.exphem.2014.01.011
Yuan N, Song L, Lin W, Cao Y, Xu F, Liu S, Zhang A, Wang Z, Li X, Fang Y, Zhang H, Zhao W, Hu S, Wang J, Zhang S (2015) Autophagy collaborates with ubiquitination to downregulate oncoprotein E2A/Pbx1 in B-cell acute lymphoblastic leukemia. Blood Cancer J 5(1):e274. doi:10.1038/bcj.2014.96
Zhang S, Song L, Yuan N, Lin W, Cao Y, Xu F, Fang Y, Wang Z, Li X, Wang Z, Cai J, Zhao W, Hu S, Chen S, Mao X, Wang J (2014) Autophagy collaborates with ubiquitination to down-regulate oncoprotein E2A/Pbx1 in B cell acute lymphoblastic leukemia. Blood 124(21):5276
Fransecky L, Mochmann LH, Baldus CD (2015) Outlook on PI3K/AKT/mTOR inhibition in acute leukemia. Mol Cell Ther 3(2):1–17. doi:10.1186/s40591-015-0040-8
Duque-Afonso J, Feng J, Scherer F, Lin C-H, Wong SHK, Wang Z, Iwasaki M, Cleary ML (2015) Comparative genomics reveals multistep pathogenesis of E2A-PBX1 acute lymphoblastic leukemia. J Clin Investig 125(9):3667–3680. doi:10.1172/JCI81158
Duque-Afonso J, Lin C-H, Han K, Wei MC, Feng J, Kurzer JH, Schneidawind C, Wong SH-K, Bassik MC, Cleary ML (2016) E2A-PBX1 remodels oncogenic signaling networks in B-cell precursor acute lymphoid leukemia. Cancer Res 76(23):6937–6949. doi:10.1158/0008-5472.CAN-16-1899
Hempel WM, Schatzman RC, DeFranco AL (1992) Tyrosine phosphorylation of phospholipase C-gamma 2 upon cross-linking of membrane Ig on murine B lymphocytes. J Immunol 148(10):3021–3027
Duy C, Yu JJ, Nahar R, Swaminathan S, Kweon S-M, Polo JM, Valls E, Klemm L, Shojaee S, Cerchietti L, Schuh W, Jäck H-M, Hurtz C, Ramezani-Rad P, Herzog S, Jumaa H, Koeffler HP, de Alborán IM, Melnick AM, Ye BH, Müschen M (2010) BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med 207(6):1209–1221. doi:10.1084/jem.20091299
Müschen M (2015) Rationale for targeting the pre–B-cell receptor signaling pathway in acute lymphoblastic leukemia. Blood 125(24):3688–3693. doi:10.1182/blood-2015-01-567842
Geng H, Hurtz C, Lenz Kyle B, Chen Z, Baumjohann D, Thompson S, Goloviznina NA, Chen W-Y, Huan J, LaTocha D, Ballabio E, Xiao G, Lee J-W, Deucher A, Qi Z, Park E, Huang C, Nahar R, Kweon S-M, Shojaee S, Chan Lai N, Yu J, Kornblau Steven M, Bijl Janetta J, Ye BH, Mark Ansel K, Paietta E, Melnick A, Hunger Stephen P, Kurre P, Tyner Jeffrey W, Loh Mignon L, Roeder Robert G, Druker Brian J, Burger JA, Milne Thomas A, Chang Bill H, Müschen M (2015) Self-enforcing feedback activation between BCL6 and pre-B Cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer Cell 27(3):409–425. doi:10.1016/j.ccell.2015.02.003
Eldfors S, Kuusanmaki H, Kontro M, Majumder MM, Parsons A, Edgren H, Pemovska T, Kallioniemi O, Wennerberg K, Gokbuget N, Burmeister T, Porkka K, Heckman CA (2017) Idelalisib sensitivity and mechanisms of disease progression in relapsed TCF3-PBX1 acute lymphoblastic leukemia. Leukemia 31(1):51–57. doi:10.1038/leu.2016.202
Ramadani F, Bolland DJ, Garcon F, Emery JL, Vanhaesebroeck B, Corcoran AE, Okkenhaug K (2010) The PI3K isoforms p110α and p110δ are essential for pre-B cell receptor signaling and B cell development. Sci Signal 3(134):ra60. doi:10.1126/scisignal.2001104
Hartsink-Segers SA, Zwaan CM, Exalto C, Luijendijk MWJ, Calvert VS, Petricoin EF, Evans WE, Reinhardt D, de Haas V, Hedtjarn M, Hansen BR, Koch T, Caron HN, Pieters R, Den Boer ML (2013) Aurora kinases in childhood acute leukemia: the promise of aurora B as therapeutic target. Leukemia 27(3):560–568. doi:10.1038/leu.2012.256
Hans F, Dimitrov S (2001) Histone H3 phosphorylation and cell division. Oncogene 20(24):7
Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P, Barrientos JC, Zelenetz AD, Kipps TJ, Flinn I, Ghia P, Eradat H, Ervin T, Lamanna N, Coiffier B, Pettitt AR, Ma S, Stilgenbauer S, Cramer P, Aiello M, Johnson DM, Miller LL, Li D, Jahn TM, Dansey RD, Hallek M, O’Brien SM (2014) Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med 370(11):997–1007. doi:10.1056/NEJMoa1315226
Kurzrock R, Kantarjian HM, Druker BJ, Talpaz M (2003) Philadelphia chromosomepositive leukemias: from basic mechanisms to molecular therapeutics. Ann Intern Med 138(10):819–830. doi:10.7326/0003-4819-138-10-200305200-00010
Savage DG, Szydlo R, Goldman JM (1997) Clinical features at diagnosis in 430 patients with chronic myeloid leukaemia seen at a referral centre over a 16-year period. Br J Haematol 96:111–116. doi:10.1046/j.1365-2141.1997.d01-1982.x
Kantarjian HM, Keating MJ, Talpaz M, Walters RS, Smith TL, Cork A, McCredie KB, Freireich EJ (1987) Chronic myelogenous leukemia in blast crisis. Am J Med 83(3):445–454. doi:10.1016/0002-9343(87)90754-6
Zhao X, Ghaffari S, Lodish H, Malashkevich VN, Kim PS (2002) Structure of the Bcr-Abl oncoprotein oligomerization domain. Nat Struct Mol Biol 9(2):117–120. doi:10.1038/nsb747
Pane F, Intrieri M, Quintarelli C, Izzo B, Muccioli GC, Salvatore F (2002) BCR/ABL genes and leukemic phenotype: from molecular mechanisms to clinical correlations. Oncogene 21(56):8652–8667. doi:10.1038/sj.onc.1206094
Zou X, Calame K (1999) Signaling pathways activated by oncogenic forms of Abl tyrosine kinase. J Biol Chem 274(26):18141–18144. doi:10.1074/jbc.274.26.18141
Danial NN, Rothman P (2000) JAK-STAT signaling activated by Abl oncogenes. Oncogene 19(21):2523–2531. doi:10.1038/sj.onc.1203484
Li S, Ilaria RL, Million RP, Daley GQ, Van Etten RA (1999) The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med 189(9):1399–1412. doi:10.1084/jem.189.9.1399
Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MWN, Silver RT, Goldman JM, Stone RM, Cervantes F, Hochhaus A, Powell BL, Gabrilove JL, Rousselot P, Reiffers J, Cornelissen JJ, Hughes T, Agis H, Fischer T, Verhoef G, Shepherd J, Saglio G, Gratwohl A, Nielsen JL, Radich JP, Simonsson B, Taylor K, Baccarani M, So C, Letvak L, Larson RA (2006) Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med 355(23):2408–2417. doi:10.1056/NEJMoa062867
Ottmann OG, Druker BJ, Sawyers CL, Goldman JM, Reiffers J, Silver RT, Tura S, Fischer T, Deininger MW, Schiffer CA, Baccarani M, Gratwohl A, Hochhaus A, Hoelzer D, Fernandes-Reese S, Gathmann I, Capdeville R, Brien SG (2002) A phase 2 study of imatinib in patients with relapsed or refractory Philadelphia chromosome-positive acute lymphoid leukemias. Blood 100(6):1965. doi:10.1182/blood-2001-12-0181
Ravandi F, Brien S, Thomas D, Faderl S, Jones D, Garris R, Dara S, Jorgensen J, Kebriaei P, Champlin R, Borthakur G, Burger J, Ferrajoli A, Garcia-Manero G, Wierda W, Cortes J, Kantarjian H (2010) First report of phase 2 study of dasatinib with hyper-CVAD for the frontline treatment of patients with Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia. Blood 116(12):2070. doi:10.1182/blood-2009-12-261586
Fielding AK, Rowe JM, Buck G, Foroni L, Gerrard G, Litzow MR, Lazarus H, Luger SM, Marks DI, McMillan AK, Moorman AV, Patel B, Paietta E, Tallman MS, Goldstone AH (2014) UKALLXII/ECOG2993: addition of imatinib to a standard treatment regimen enhances long-term outcomes in Philadelphia positive acute lymphoblastic leukemia. Blood 123(6):843. doi:10.1182/blood-2013-09-529008
Schultz KR, Carroll A, Heerema NA, Bowman WP, Aledo A, Slayton WB, Sather H, Devidas M, Zheng HW, Davies SM, Gaynon PS, Trigg M, Rutledge R, Jorstad D, Winick N, Borowitz MJ, Hunger SP, Carroll WL, Camitta B (2014) Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group Study AALL0031. Leukemia 28(7):1467–1471. doi:10.1038/leu.2014.30
Biondi A, Schrappe M, De Lorenzo P, Castor A, Lucchini G, Gandemer V, Pieters R, Stary J, Escherich G, Campbell M, Li C-K, Vora A, Aricò M, Röttgers S, Saha V, Valsecchi MG (2012) Imatinib after induction for treatment of children and adolescents with Philadelphia-chromosome-positive acute lymphoblastic leukaemia (EsPhALL): a randomised, open-label, intergroup study. Lancet Oncol 13(9):936–945. doi:10.1016/S1470-2045(12)70377-7
Bardini M, Spinelli R, Bungaro S, Mangano E, Corral L, Cifola I, Fazio G, Giordan M, Basso G, De Rossi G, Biondi A, Battaglia C, Cazzaniga G (2009) DNA copy-number abnormalities do not occur in infant ALL with t(4;11)/MLL-AF4. Leukemia 24(1):169–176. doi:10.1038/leu.2009.203
Wylie AA, Schoepfer J, Jahnke W, Cowan-Jacob SW, Loo A, Furet P, Marzinzik AL, Pelle X, Donovan J, Zhu W, Buonamici S, Hassan AQ, Lombardo F, Iyer V, Palmer M, Berellini G, Dodd S, Thohan S, Bitter H, Branford S, Ross DM, Hughes TP, Petruzzelli L, Vanasse KG, Warmuth M, Hofmann F, Keen NJ, Sellers WR (2017) The allosteric inhibitor ABL001 enables dual targeting of BCR–ABL1. Nature 543(7647):733–737. doi:10.1038/nature21702
Weisberg E, Manley P, Mestan J, Cowan-Jacob S, Ray A, Griffin JD (2006) AMN107 (nilotinib): a novel and selective inhibitor of BCR-ABL. Br J Cancer 94(12):1765–1769. doi:10.1038/sj.bjc.6603170
Perez-Andreu V, Roberts KG, Xu H, Smith C, Zhang H, Yang W, Harvey RC, Payne-Turner D, Devidas M, Cheng IM, Carroll WL, Heerema NA, Carroll AJ, Raetz EA, Gastier-Foster JM, Marcucci G, Bloomfield CD, Mrózek K, Kohlschmidt J, Stock W, Kornblau SM, Konopleva M, Paietta E, Rowe JM, Luger SM, Tallman MS, Dean M, Burchard EG, Torgerson DG, Yue F, Wang Y, Pui C-H, Jeha S, Relling MV, Evans WE, Gerhard DS, Loh ML, Willman CL, Hunger SP, Mullighan CG, Yang JJ (2015) A genome-wide association study of susceptibility to acute lymphoblastic leukemia in adolescents and young adults. Blood 125(4):680–686. doi:10.1182/blood-2014-09-595744
Perez-Andreu V, Roberts KG, Harvey RC, Yang W, Cheng C, Pei D, Xu H, Gastier-Foster J, Lim JY-S, Chen IM, Fan Y, Devidas M, Borowitz MJ, Smith C, Neale G, Burchard EG, Torgerson DG, Klussmann FA, Villagran CRN, Winick NJ, Camitta BM, Raetz E, Wood B, Yue F, Carroll WL, Larsen E, Bowman WP, Loh ML, Dean M, Bhojwani D, Pui C-H, Evans WE, Relling MV, Hunger SP, Willman CL, Mullighan CG, Yang JJ (2013) Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat Genet 45(12):1494–1498. doi:10.1038/ng.2803
Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui C-H, Relling MV, Shurtleff SA, Downing JR (2008) BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453(7191):110–114. doi:10.1038/nature06866
Witkowski MT, Hu Y, Roberts KG, Boer JM, McKenzie MD, Liu GJ, Le Grice OD, Tremblay CS, Ghisi M, Willson TA, Horstmann MA, Aifantis I, Cimmino L, Frietze S, den Boer ML, Mullighan CG, Smyth GK, Dickins RA (2017) Conserved IKAROS-regulated genes associated with B-progenitor acute lymphoblastic leukemia outcome. J Exp Med 214(6):1–19. doi:10.1084/jem.20160048
Li W, Dreazen O, Kloetzer WS, Gale R, Arlinghaus R (1989) Characterization of bcr gene products in hematopoietic cells. Oncogene 4(2):127–138
Maru Y, Witte ON (1991) The BCR gene encodes a novel serine/threonine kinase activity within a single exon. Cell 67(3):459–468. doi:10.1016/0092-8674(91)90521-Y
Maru Y, Peters KL, Afar DE, Shibuya M, Witte ON, Smithgall TE (1995) Tyrosine phosphorylation of BCR by FPS/FES protein-tyrosine kinases induces association of BCR with GRB-2/SOS. Mol Cell Biol 15(2):835–842
Gu S, Chan WW, Mohi G, Rosenbaum J, Sayad A, Lu Z, Virtanen C, Li S, Neel BG, Van Etten RA (2016) Distinct GAB2 signaling pathways are essential for myeloid and lymphoid transformation and leukemogenesis by BCR-ABL1. Blood 127(14):1803–1813. doi:10.1182/blood-2015-06-653006
Liu J, Wu Y, Ma GZ, Lu D, Haataja L, Heisterkamp N, Groffen J, Arlinghaus RB (1996) Inhibition of Bcr serine kinase by tyrosine phosphorylation. Mol Cell Biol 16(3):998–1005. doi:10.1128/MCB.16.3.998
Schwartzberg PL, Stall AM, Hardin JD, Bowdish KS, Humaran T, Boast S, Harbison ML, Robertson EJ, Goff SP (1991) Mice homozygous for the abl m1 mutation show poor viability and depletion of selected B and T cell populations. Cell 65(7):1165–1175. doi:10.1016/0092-8674(91)90012-N
Rosti V, Bergamaschi G, Lucotti C, Danova M, Carlo-Stella C, Locatelli F, Tonon L, Mazzini G, Cazzola M (1995) Oligodeoxynucleotides antisense to c-abl specifically inhibit entry into S-phase of CD34+ hematopoietic cells and their differentiation to granulocyte-macrophage progenitors. Blood 86(9):3387–3393
Tao W, Leng X, Chakraborty SN, Ma H, Arlinghaus RB (2014) c-Abl activates janus kinase 2 in normal hematopoietic cells. J Biol Chem 289(31):21463–21472. doi:10.1074/jbc.M114.554501
Heisterkamp N, Jenster G, ten Hoeve J, Zovich D, Pattengale PK, Groffen J (1990) Acute leukaemia in bcr/abl transgenic mice. Nature 344(6263):251–253. doi:10.1038/344251a0
Voncken JW, Morris C, Pattengale P, Dennert G, Kikly C, Groffen J, Heisterkamp N (1992) Clonal development and karyotype evolution during leukemogenesis of BCR/ABL transgenic mice. Blood 79(4):1029–1036
Heisterkamp N, Jenster G, Kioussis D, Pattengale PK, Groffen J (1991) Human bcr-abl gene has a lethal effect on embryogenesis. Transgenic Res 1(1):45–53. doi:10.1007/BF02512996
Smith MR, DeGudicibus SJ, Stacey DW (1986) Requirement for c-ras proteins during viral oncogene transformation. Nature 320(6062):540–543. doi:10.1038/320540a0
Hancock JF (2003) Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol 4(5):373–385. doi:10.1038/nrm1105
Kato K, Cox AD, Hisaka MM, Graham SM, Buss JE, Der CJ (1992) Isoprenoid addition to Ras protein is the critical modification for its membrane association and transforming activity. Proc Natl Acad Sci 89(14):6403–6407
Reichert A, Heisterkamp N, Daley GQ, Groffen J (2001) Treatment of Bcr/Abl-positive acute lymphoblastic leukemia in P190 transgenic mice with the farnesyl transferase inhibitor SCH66336. Blood 97(5):1399–1403. doi:10.1182/blood.V97.5.1399
Skorski T, Kanakaraj P, Nieborowska-Skorska M, Ratajczak MZ, Wen SC, Zon G, Gewirtz AM, Perussia B, Calabretta B (1995) Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood 86(2):726–736
Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi JK, Trotta R, Wlodarski P, Perrotti D, Chan TO, Wasik MA, Tsichlis PN, Calabretta B (1997) Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J 16(20):6151–6161. doi:10.1093/emboj/16.20.6151
Kharas MG, Janes MR, Scarfone VM, Lilly MB, Knight ZA, Shokat KM, Fruman DA (2008) Ablation of PI3K blocks BCR-ABL leukemogenesis in mice, and a dual PI3K/mTOR inhibitor prevents expansion of human BCR-ABL+ leukemia cells. J Clin Investig 118(9):3038–3050. doi:10.1172/JCI33337
Raynaud FI, Eccles S, Clarke PA, Hayes A, Nutley B, Alix S, Henley A, Di-Stefano F, Ahmad Z, Guillard S, Bjerke LM, Kelland L, Valenti M, Patterson L, Gowan S, de Haven Brandon A, Hayakawa M, Kaizawa H, Koizumi T, Ohishi T, Patel S, Saghir N, Parker P, Waterfield M, Workman P (2007) Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res 67(12):5840–5850. doi:10.1158/0008-5472.CAN-06-4615
Henry CJ, Marusyk A, Zaberezhnyy V, Adane B, DeGregori J (2010) Declining lymphoid progenitor fitness promotes aging-associated leukemogenesis. Proc Natl Acad Sci 107(50):21713–21718. doi:10.1073/pnas.1005486107
Notta F, Mullighan CG, Wang JCY, Poeppl A, Doulatov S, Phillips LA, Ma J, Minden MD, Downing JR, Dick JE (2011) Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature 469(7330):362–367. doi:10.1038/nature09733
Waldron T, De Dominici M, Soliera AR, Audia A, Iacobucci I, Lonetti A, Martinelli G, Zhang Y, Martinez R, Hyslop T, Bender TP, Calabretta B (2012) c-Myb and its target Bmi1 are required for p190BCR/ABL leukemogenesis in mouse and human cells. Leukemia 26(4):644–653. doi:10.1038/leu.2011.264
Steenbergen EJ, Verhagen OJ, van Leeuwen EF, von dem Borne AE, van der Schoot CE (1993) Distinct ongoing Ig heavy chain rearrangement processes in childhood B- precursor acute lymphoblastic leukemia. Blood 82(2):581–589
Gawad C, Pepin F, Carlton VEH, Klinger M, Logan AC, Miklos DB, Faham M, Dahl G, Lacayo N (2012) Massive evolution of the immunoglobulin heavy chain locus in children with B precursor acute lymphoblastic leukemia. Blood 120(22):4407–4417. doi:10.1182/blood-2012-05-429811
Kordes U, Krappmann D, Heissmeyer V, Ludwig W, Scheidereit C (2000) Transcription factor NF-kappaB is constitutively activated in acute lymphoblastic leukemia cells. Leukemia 14(3):399–402
Ochodnicka-Mackovicova K, Bahjat M, Bloedjes TA, Maas C, de Bruin AM, Bende RJ, van Noesel CJM, Guikema JEJ (2015) NF-κB and AKT signaling prevent DNA damage in transformed pre-B cells by suppressing RAG1/2 expression and activity. Blood 126(11):1324–1335. doi:10.1182/blood-2015-01-621623
Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, Meyer JD, Davison BL (1994) Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med 180(5):1955–1960. doi:10.1084/jem.180.5.1955
Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, te Kronnie G, Cario G, Cazzaniga G, Kulozik AE, Stanulla M, Schrappe M, Biondi A, Basso G, Bercovich D, Muckenthaler MU, Izraeli S (2011) Gain-of-function mutations in interleukin-7 receptor-α (IL7R) in childhood acute lymphoblastic leukemias. J Exp Med 208(5):901–908. doi:10.1084/jem.20110580
Ilaria RL, Van Etten RA (1996) P210 and P190BCR/ABL induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem 271(49):31704–31710. doi:10.1074/jbc.271.49.31704
Chai SK, Nichols GL, Rothman P (1997) Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J Immunol 159(10):4720–4728
Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, Tasian SK, Loh ML, Su X, Liu W, Devidas M, Atlas SR, Chen I-M, Clifford RJ, Gerhard DS, Carroll WL, Reaman GH, Smith M, Downing JR, Hunger SP, Willman CL (2009) JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci 106(23):9414–9418. doi:10.1073/pnas.0811761106
Miyamoto N, Sugita K, Goi K, Inukai T, Iijima K, Tezuka T, Kojika S, Nakamura M, Kagami K, Nakazawa S (2001) The JAK2 inhibitor AG490 predominantly abrogates the growth of human B-precursor leukemic cells with 11q23 translocation or Philadelphia chromosome. Leukemia 15(11):1758–1768
Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJCM, Beverloo HB, Van der Spek PJ, Escherich G, Horstmann MA, Janka-Schaub GE, Kamps WA, Evans WE, Pieters R (2009) A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol 10(2):125–134. doi:10.1016/S1470-2045(08)70339-5
Boer JM, Koenders JE, van der Holt B, Exalto C, Sanders MA, Cornelissen JJ, Valk PJM, den Boer ML, Rijneveld AW (2015) Expression profiling of adult acute lymphoblastic leukemia identifies a BCR-ABL1-like subgroup characterized by high non-response and relapse rates. Haematologica 100(7):e261–e264. doi:10.3324/haematol.2014.117424
Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang Y-L, Pei D, McCastlain K, Ding L, Lu C, Song G, Ma J, Becksfort J, Rusch M, Chen S-C, Easton J, Cheng J, Boggs K, Santiago-Morales N, Iacobucci I, Fulton RS, Wen J, Valentine M, Cheng C, Paugh SW, Devidas M, Chen IM, Reshmi S, Smith A, Hedlund E, Gupta P, Nagahawatte P, Wu G, Chen X, Yergeau D, Vadodaria B, Mulder H, Winick NJ, Larsen EC, Carroll WL, Heerema NA, Carroll AJ, Grayson G, Tasian SK, Moore AS, Keller F, Frei-Jones M, Whitlock JA, Raetz EA, White DL, Hughes TP, Guidry Auvil JM, Smith MA, Marcucci G, Bloomfield CD, Mrózek K, Kohlschmidt J, Stock W, Kornblau SM, Konopleva M, Paietta E, Pui C-H, Jeha S, Relling MV, Evans WE, Gerhard DS, Gastier-Foster JM, Mardis E, Wilson RK, Loh ML, Downing JR, Hunger SP, Willman CL, Zhang J, Mullighan CG (2014) Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 371(11):1005–1015. doi:10.1056/NEJMoa1403088
Roberts Kathryn G, Morin Ryan D, Zhang J, Hirst M, Zhao Y, Su X, Chen S-C, Payne-Turner D, Churchman ML, Harvey Richard C, Chen X, Kasap C, Yan C, Becksfort J, Finney Richard P, Teachey David T, Maude Shannon L, Tse K, Moore R, Jones S, Mungall K, Birol I, Edmonson Michael N, Hu Y, Buetow Kenneth E, Chen IM, Carroll William L, Wei L, Ma J, Kleppe M, Levine Ross L, Garcia-Manero G, Larsen E, Shah Neil P, Devidas M, Reaman G, Smith M, Paugh Steven W, Evans William E, Grupp Stephan A, Jeha S, Pui C-H, Gerhard Daniela S, Downing James R, Willman Cheryl L, Loh M, Hunger Stephen P, Marra Marco A, Mullighan Charles G (2012) Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell 22(2):153–166. doi:10.1016/j.ccr.2012.06.005
Lengline E, Beldjord K, Dombret H, Soulier J, Boissel N, Clappier E (2013) Successful tyrosine kinase inhibitor therapy in a refractory B-cell precursor acute lymphoblastic leukemia with EBF1-PDGFRB fusion. Haematologica 98(11):e146–e148. doi:10.3324/haematol.2013.095372
Maude SL, Tasian SK, Vincent T, Hall JW, Sheen C, Roberts KG, Seif AE, Barrett DM, Chen IM, Collins JR, Mullighan CG, Hunger SP, Harvey RC, Willman CL, Fridman JS, Loh ML, Grupp SA, Teachey DT (2012) Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood 120(17):3510–3518. doi:10.1182/blood-2012-03-415448
Loh ML, Tasian SK, Rabin KR, Brown P, Magoon D, Reid JM, Chen X, Ahern CH, Weigel BJ, Blaney SM (2015) A phase 1 dosing study of ruxolitinib in children with relapsed or refractory solid tumors, leukemias, or myeloproliferative neoplasms: a Children’s Oncology Group phase 1 consortium study (ADVL1011). Pediatr Blood Cancer 62(10):1717–1724. doi:10.1002/pbc.25575
Maia AT, Tussiwand R, Cazzaniga G, Rebulla P, Colman S, Biondi A, Greaves M (2004) Identification of preleukemic precursors of hyperdiploid acute lymphoblastic leukemia in cord blood. Genes Chromosom Cancer 40(1):38–43. doi:10.1002/gcc.20010
Ito C, M-a Kumagai, Manabe A, Coustan-Smith E, Raimondi SC, Behm FG, Murti KG, Rubnitz JE, Pui C-H, Campana D (1999) Hyperdiploid acute lymphoblastic leukemia with 51 to 65 chromosomes: a distinct biological entity with a marked propensity to undergo apoptosis. Blood 93(1):315–320
Zhang Y, Lu J, van den Berghe J, Lee S-H (2002) Increased incidence of spontaneous apoptosis in the bone marrow of hyperdiploid childhood acute lymphoblastic leukemia. Exp Hematol 30(4):333–339. doi:10.1016/S0301-472X(02)00771-3
Armstrong SA, Mabon ME, Silverman LB, Li A, Gribben JG, Fox EA, Sallan SE, Korsmeyer SJ (2004) FLT3 mutations in childhood acute lymphoblastic leukemia. Blood 103(9):3544. doi:10.1182/blood-2003-07-2441
Wiemels JL, Zhang Y, Chang J, Zheng S, Metayer C, Zhang L, Smith MT, Ma X, Selvin S, Buffler PA, Wiencke JK (2005) RAS mutation is associated with hyperdiploidy and parental characteristics in pediatric acute lymphoblastic leukemia. Leukemia 19(3):415–419. doi:10.1038/sj.leu.2403641
Tartaglia M, Martinelli S, Cazzaniga G, Cordeddu V, Iavarone I, Spinelli M, Palmi C, Carta C, Pession A, Aricò M, Masera G, Basso G, Sorcini M, Gelb BD, Biondi A (2004) Genetic evidence for lineage-related and differentiation stage–related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood 104(2):307–313. doi:10.1182/blood-2003-11-3876
Paulsson K, Horvat A, Strömbeck B, Nilsson F, Heldrup J, Behrendtz M, Forestier E, Andersson A, Fioretos T, Johansson B (2008) Mutations of FLT3, NRAS, KRAS, and PTPN11 are frequent and possibly mutually exclusive in high hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosom Cancer 47(1):26–33. doi:10.1002/gcc.20502
Paulsson K, Forestier E, Lilljebjörn H, Heldrup J, Behrendtz M, Young BD, Johansson B (2010) Genetic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Proc Natl Acad Sci 107(50):21719–21724. doi:10.1073/pnas.1006981107
Safavi S, Paulsson K (2017) Near-haploid and low-hypodiploid acute lymphoblastic leukemia: two distinct subtypes with consistently poor prognosis. Blood 129(4):420–423. doi:10.1182/blood-2016-10-743765
Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, Payne-Turner D, Churchman M, Andersson A, Chen S-C, McCastlain K, Becksfort J, Ma J, Wu G, Patel SN, Heatley SL, Phillips LA, Song G, Easton J, Parker M, Chen X, Rusch M, Boggs K, Vadodaria B, Hedlund E, Drenberg C, Baker S, Pei D, Cheng C, Huether R, Lu C, Fulton RS, Fulton LL, Tabib Y, Dooling DJ, Ochoa K, Minden M, Lewis ID, To LB, Marlton P, Roberts AW, Raca G, Stock W, Neale G, Drexler HG, Dickins RA, Ellison DW, Shurtleff SA, Pui C-H, Ribeiro RC, Devidas M, Carroll AJ, Heerema NA, Wood B, Borowitz MJ, Gastier-Foster JM, Raimondi SC, Mardis ER, Wilson RK, Downing JR, Hunger SP, Loh ML, Mullighan CG (2013) The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 45(3):242–252. doi:10.1038/ng.2532
Harris MB, Shuster JJ, Carroll A, Look AT, Borowitz MJ, Crist WM, Nitschke R, Pullen J, Steuber CP, Land VJ (1992) Trisomy of leukemic cell chromosomes 4 and 10 identifies children with B-progenitor cell acute lymphoblastic leukemia with a very low risk of treatment failure: a Pediatric Oncology Group study. Blood 79(12):3316
Berger R (1997) Acute lymphoblastic leukemia and chromosome 21. Cancer Genet Cytogenet 94(1):8–12. doi:10.1016/S0165-4608(96)00351-2
Mateos MK, Barbaric D, Byatt S-A, Sutton R, Marshall GM (2015) Down syndrome and leukemia: insights into leukemogenesis and translational targets. Transl Pediatr 4(2):76–92. doi:10.3978/j.issn.2224-4336.2015.03.03
Harewood L, Robinson H, Harris R, Al-Obaidi MJ, Jalali GR, Martineau M, Moorman AV, Sumption N, Richards S, Mitchell C, Harrison CJ (2003) Amplification of AML1 on a duplicated chromosome 21 in acute lymphoblastic leukemia: a study of 20 cases. Leukemia 17(3):547–553. doi:10.1038/sj.leu.2402849
Heerema NA, Carroll AJ, Devidas M, Loh ML, Borowitz MJ, Gastier-Foster JM, Larsen EC, Mattano LA, Maloney KW, Willman CL, Wood BL, Winick NJ, Carroll WL, Hunger SP, Raetz EA (2013) Intrachromosomal amplification of chromosome 21 is associated with inferior outcomes in children with acute lymphoblastic leukemia treated in contemporary standard-risk children’s oncology group studies: a report from the Children’s Oncology Group. J Clin Oncol 31(27):3397–3402. doi:10.1200/JCO.2013.49.1308
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Malouf, C., Ottersbach, K. Molecular processes involved in B cell acute lymphoblastic leukaemia. Cell. Mol. Life Sci. 75, 417–446 (2018). https://doi.org/10.1007/s00018-017-2620-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-017-2620-z