Abstract
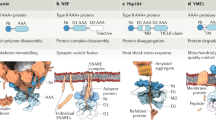
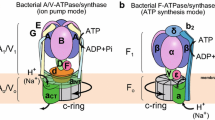
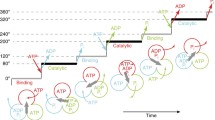
ATPases Associated with various cellular Activities (AAA+ ATPases) are molecular motors that use the energy of ATP binding and hydrolysis to remodel their target macromolecules. The majority of these ATPases form ring-shaped hexamers in which the active sites are located at the interfaces between neighboring subunits. Structural changes initiate in an active site and propagate to distant motor parts that interface and reshape the target macromolecules, thereby performing mechanical work. During the functioning cycle, the AAA+ motor transits through multiple distinct states. Ring architecture and placement of the catalytic sites at the intersubunit interfaces allow for a unique level of coordination among subunits of the motor. This in turn results in conformational differences among subunits and overall asymmetry of the motor ring as it functions. To date, a large amount of structural information has been gathered for different AAA+ motors, but even for the most characterized of them only a few structural states are known and the full mechanistic cycle cannot be yet reconstructed. Therefore, the first part of this work will provide a broad overview of what arrangements of AAA+ subunits have been structurally observed focusing on diversity of ATPase oligomeric ensembles and heterogeneity within the ensembles. The second part of this review will concentrate on methods that assess structural and functional heterogeneity among subunits of AAA+ motors, thus bringing us closer to understanding the mechanism of these fascinating molecular motors.





Similar content being viewed by others
References
Neuwald AF, Aravind L, Spouge JL, Koonin EV (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9:27–43
Ogura T, Wilkinson AJ (2001) AAA+ superfamily ATPases: common structure–diverse function. Genes Cells 6:575–597
Maurizi MR, Li CC (2001) AAA proteins: in search of a common molecular basis. International meeting on cellular functions of AAA proteins. EMBO Rep 2:980–985
Lupas AN, Martin J (2002) AAA proteins. Curr Opin Struct Biol 12:746–753
Wang J (2004) Nucleotide-dependent domain motions within rings of the RecA/AAA(+) superfamily. J Struct Biol 148:259–267
Ammelburg M, Frickey T, Lupas AN (2006) Classification of AAA+ proteins. J Struct Biol 156:2–11
Hanson PI, Whiteheart SW (2005) AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6:519–529
Iyer LM, Leipe DD, Koonin EV, Aravind L (2004) Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol 146:11–31
White SR, Lauring B (2007) AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic 8:1657–1667
Barthelme D, Jauregui R, Sauer RT (2015) An ALS disease mutation in Cdc48/p97 impairs 20S proteasome binding and proteolytic communication. Protein Sci 24:1521–1527
Xia D, Tang WK, Ye Y (2016) Structure and function of the AAA+ ATPASE p97/Cdc48p. Gene 583:64–77
Hafezparast M, Klocke R, Ruhrberg C, Marquardt A et al (2003) Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science 300:808–812
Eschbach J, Dupuis L (2011) Cytoplasmic dynein in neurodegeneration. Pharmacol Ther 130:348–363
Chen XJ, Xu H, Cooper HM, Liu Y (2014) Cytoplasmic dynein: a key player in neurodegenerative and neurodevelopmental diseases. Sci China Life Sci 57:372–377
Boyaci H, Shah T, Hurley A, Kokona B et al (2016) Structure, regulation, and inhibition of the quorum-sensing signal integrator LuxO. PLoS Biol 14:e1002464
Erzberger JP, Berger JM (2006) Evolutionary relationships and structural mechanisms of AAA plus proteins. Annu Rev Biophys Biomol Struct 35:93–114
Thomsen ND, Berger JM (2008) Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases. Mol Microbiol 69:1071–1090
Frickey T, Lupas AN (2004) Phylogenetic analysis of AAA proteins. J Struct Biol 146:2–10
Iyer LM, Makarova KS, Koonin EV, Aravind L (2004) Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res 32:5260–5279
Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1:945–951
Snider J, Houry WA (2008) AAA+ proteins: diversity in function, similarity in structure. Biochem Soc Trans 36:72–77
Wendler P, Ciniawsky S, Kock M, Kube S (2012) Structure and function of the AAA+ nucleotide binding pocket. Biochim Biophys Acta 1823:2–14
Ogura T, Whiteheart SW, Wilkinson AJ (2004) Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol 146:106–112
Zhang X, Wigley DB (2008) The ‘glutamate switch’ provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat Struct Mol Biol 15:1223–1227
Lee SY, De La Torre A, Yan D, Kustu S et al (2003) Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev 17:2552–2563
Park E, Rho YM, Koh OJ, Ahn SW et al (2005) Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J Biol Chem 280:22892–22898
Enemark EJ, Joshua-Tor L (2006) Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442:270–275
Sauer RT, Baker TA (2011) AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem 80:587–612
Arias-Palomo E, Berger JM (2015) An atypical AAA+ ATPase assembly controls efficient transposition through DNA remodeling and transposase recruitment. Cell 162:860–871
Kon T, Sutoh K, Kurisu G (2011) X-ray structure of a functional full-length dynein motor domain. Nat Struct Mol Biol 18:638–642
Carter AP, Cho C, Jin L, Vale RD (2011) Crystal structure of the dynein motor domain. Science 331:1159–1165
Chen B, Sysoeva TA, Chowdhury S, Guo L et al (2010) Engagement of arginine finger to ATP triggers large conformational changes in NtrC1 AAA+ ATPase for remodeling bacterial RNA polymerase. Structure 18:1420–1430
Batchelor JD, Sterling HJ, Hong E, Williams ER et al (2009) Receiver domains control the active-state stoichiometry of Aquifex aeolicus sigma54 activator NtrC4, as revealed by electrospray ionization mass spectrometry. J Mol Biol 393:634–643
Dey S, Biswas M, Sen U, Dasgupta J (2015) Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC. J Biol Chem 290:8734–8747
Rohrwild M, Pfeifer G, Santarius U, Muller SA et al (1997) The ATP-dependent HslVU protease from Escherichia coli is a four-ring structure resembling the proteasome. Nat Struct Biol 4:133–139
Toth EA, Li Y, Sawaya MR, Cheng Y et al (2003) The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol Cell 12:1113–1123
Singleton MR, Sawaya MR, Ellenberger T, Wigley DB (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101:589–600
Ziebarth TD, Gonzalez-Soltero R, Makowska-Grzyska MM, Nunez-Ramirez R et al (2010) Dynamic effects of cofactors and DNA on the oligomeric state of human mitochondrial DNA helicase. J Biol Chem 285:14639–14647
Yu X, VanLoock MS, Poplawski A, Kelman Z et al (2002) The Methanobacterium thermoautotrophicum MCM protein can form heptameric rings. EMBO Rep 3:792–797
Gomez-Llorente Y, Fletcher RJ, Chen XS, Carazo JM et al (2005) Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J Biol Chem 280:40909–40915
Kim KI, Cheong GW, Park SC, Ha JS et al (2000) Heptameric ring structure of the heat-shock protein ClpB, a protein-activated ATPase in Escherichia coli. J Mol Biol 303:655–666
Lee S, Sowa ME, Watanabe YH, Sigler PB et al (2003) The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115:229–240
Watanabe YH, Takano M, Yoshida M (2005) ATP binding to nucleotide binding domain (NBD)1 of the ClpB chaperone induces motion of the long coiled-coil, stabilizes the hexamer, and activates NBD2. J Biol Chem 280:24562–24567
Akoev V, Gogol EP, Barnett ME, Zolkiewski M (2004) Nucleotide-induced switch in oligomerization of the AAA+ ATPase ClpB. Protein Sci 13:567–574
Davies JM, Brunger AT, Weis WI (2008) Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure 16:715–726
Zhang X, Shaw A, Bates PA, Newman RH et al (2000) Structure of the AAA ATPase p97. Mol Cell 6:1473–1484
Cha SS, An YJ, Lee CR, Lee HS et al (2010) Crystal structure of Lon protease: molecular architecture of gated entry to a sequestered degradation chamber. EMBO J 29:3520–3530
Stahlberg H, Kutejova E, Suda K, Wolpensinger B et al (1999) Mitochondrial Lon of Saccharomyces cerevisiae is a ring-shaped protease with seven flexible subunits. Proc Natl Acad Sci USA 96:6787–6790
Miyata T, Yamada K, Iwasaki H, Shinagawa H et al (2000) Two different oligomeric states of the RuvB branch migration motor protein as revealed by electron microscopy. J Struct Biol 131:83–89
Kazmirski SL, Podobnik M, Weitze TF, O’Donnell M et al (2004) Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex. Proc Natl Acad Sci USA 101:16750–16755
Kelch BA, Makino DL, O’Donnell M, Kuriyan J (2011) How a DNA polymerase clamp loader opens a sliding clamp. Science 334:1675–1680
Hedglin M, Kumar R, Benkovic SJ (2013) Replication clamps and clamp loaders. Cold Spring Harb Perspect Biol 5:a010165
Alam TI, Rao VB (2008) The ATPase domain of the large terminase protein, gp17, from bacteriophage T4 binds DNA: implications to the DNA packaging mechanism. J Mol Biol 376:1272–1281
Mao H, Saha M, Reyes-Aldrete E, Sherman MB et al (2016) Structural and molecular basis for coordination in a viral DNA packaging motor. Cell Rep 14:2017–2029
Moffitt JR, Chemla YR, Aathavan K, Grimes S et al (2009) Intersubunit coordination in a homomeric ring ATPase. Nature 457:446–450
Schwartz C, De Donatis GM, Fang H, Guo P (2013) The ATPase of the phi29 DNA packaging motor is a member of the hexameric AAA+ superfamily. Virology 443:20–27
Schwartz C, De Donatis GM, Zhang H, Fang H et al (2013) Revolution rather than rotation of AAA+ hexameric phi29 nanomotor for viral dsDNA packaging without coiling. Virology 443:28–39
Qi S, Pang Y, Hu Q, Liu Q et al (2010) Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141:446–457
Mansilla-Soto J, Yoon-Robarts M, Rice WJ, Arya S et al (2009) DNA structure modulates the oligomerization properties of the AAV initiator protein Rep68. PLoS Pathog 5:e1000513
Putnam CD, Clancy SB, Tsuruta H, Gonzalez S et al (2001) Structure and mechanism of the RuvB Holliday junction branch migration motor. J Mol Biol 311:297–310
Sawaya MR, Guo S, Tabor S, Richardson CC et al (1999) Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell 99:167–177
Kim DY, Kim KK (2003) Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J Biol Chem 278:50664–50670
Erzberger JP, Mott ML, Berger JM (2006) Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol 13:676–683
Rappas M, Schumacher J, Beuron F, Niwa H et al (2005) Structural insights into the activity of enhancer-binding proteins. Science 307:1972–1975
Yu RC, Hanson PI, Jahn R, Brunger AT (1998) Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat Struct Biol 5:803–811
Lenzen CU, Steinmann D, Whiteheart SW, Weis WI (1998) Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell 94:525–536
Lee S, Choi JM, Tsai FT (2007) Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol Cell 25:261–271
Lee S, Sielaff B, Lee J, Tsai FT (2010) CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc Natl Acad Sci USA 107:8135–8140
Wendler P, Shorter J, Plisson C, Cashikar AG et al (2007) Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell 131:1366–1377
Wendler P, Shorter J, Snead D, Plisson C et al (2009) Motor mechanism for protein threading through Hsp104. Mol Cell 34:81–92
Gardner BM, Chowdhury S, Lander GC, Martin A (2015) The Pex1/Pex6 complex is a heterohexameric AAA+ motor with alternating and highly coordinated subunits. J Mol Biol 427:1375–1388
Cheung KL, Huen J, Kakihara Y, Houry WA et al (2010) Alternative oligomeric states of the yeast Rvb1/Rvb2 complex induced by histidine tags. J Mol Biol 404:478–492
Nguyen VQ, Ranjan A, Stengel F, Wei D et al (2013) Molecular architecture of the ATP-dependent chromatin-remodeling complex SWR1. Cell 154:1220–1231
Champion PA, Stanley SA, Champion MM, Brown EJ et al (2006) C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 313:1632–1636
Ramsdell TL, Huppert LA, Sysoeva TA, Fortune SM et al (2015) Linked domain architectures allow for specialization of function in the FtsK/SpoIIIE ATPases of ESX secretion systems. J Mol Biol 427:1119–1132
Rosenberg OS, Dovala D, Li X, Connolly L et al (2015) Substrates control multimerization and activation of the multi-domain ATPase motor of type VII secretion. Cell 161:501–512
Ates LS, Ummels R, Commandeur S, van de Weerd R et al (2015) Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genet 11:e1005190
Glynn SE, Martin A, Nager AR, Baker TA et al (2009) Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139:744–756
Fouts ET, Yu X, Egelman EH, Botchan MR (1999) Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J Biol Chem 274:4447–4458
Bochtler M, Hartmann C, Song HK, Bourenkov GP et al (2000) The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature 403:800–805
Bieniossek C, Niederhauser B, Baumann UM (2009) The crystal structure of apo-FtsH reveals domain movements necessary for substrate unfolding and translocation. Proc Natl Acad Sci USA 106:21579–21584
Caillat C, Macheboeuf P, Wu Y, McCarthy AA et al (2015) Asymmetric ring structure of Vps4 required for ESCRT-III disassembly. Nat Commun 6:8781
Lin CC, Su SC, Su MY, Liang PH et al (2016) Structural insights into the allosteric operation of the lon AAA+ protease. Structure 24:667–675
Sanders CM, Kovalevskiy OV, Sizov D, Lebedev AA et al (2007) Papillomavirus E1 helicase assembly maintains an asymmetric state in the absence of DNA and nucleotide cofactors. Nucleic Acids Res 35:6451–6457
Itsathitphaisarn O, Wing RA, Eliason WK, Wang J et al (2012) The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 151:267–277
Cuesta I, Nunez-Ramirez R, Scheres SH, Gai D et al (2010) Conformational rearrangements of SV40 large T antigen during early replication events. J Mol Biol 397:1276–1286
Sysoeva TA, Chowdhury S, Guo L, Nixon BT (2013) Nucleotide-induced asymmetry within ATPase activator ring drives sigma54-RNAP interaction and ATP hydrolysis. Genes Dev 27:2500–2511
Zhao M, Wu S, Zhou Q, Vivona S et al (2015) Mechanistic insights into the recycling machine of the SNARE complex. Nature 518:61–67
Crampton DJ, Ohi M, Qimron U, Walz T et al (2006) Oligomeric states of bacteriophage T7 gene 4 primase/helicase. J Mol Biol 360:667–677
O’Shea VL, Berger JM (2014) Loading strategies of ring-shaped nucleic acid translocases and helicases. Curr Opin Struct Biol 25:16–24
Arias-Palomo E, O’Shea VL, Hood IV, Berger JM (2013) The bacterial DnaC helicase loader is a DnaB ring breaker. Cell 153:438–448
Blok NB, Tan D, Wang RY, Penczek PA et al (2015) Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy. Proc Natl Acad Sci USA 112:E4017–E4025
Tye BK (1999) MCM proteins in DNA replication. Annu Rev Biochem 68:649–686
Bar-Nun S, Glickman MH (2012) Proteasomal AAA-ATPases: structure and function. Biochim Biophys Acta 1823:67–82
Cho C, Vale RD (2012) The mechanism of dynein motility: insight from crystal structures of the motor domain. Biochim Biophys Acta 1823:182–191
Garbarino JE, Gibbons IR (2002) Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genom 3:18
Silvanovich A, Li MG, Serr M, Mische S et al (2003) The third P-loop domain in cytoplasmic dynein heavy chain is essential for dynein motor function and ATP-sensitive microtubule binding. Mol Biol Cell 14:1355–1365
Kon T, Nishiura M, Ohkura R, Toyoshima YY et al (2004) Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry 43:11266–11274
Cho C, Reck-Peterson SL, Vale RD (2008) Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J Biol Chem 283:25839–25845
Martin A, Baker TA, Sauer RT (2005) Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature 437:1115–1120
Joly N, Buck M (2010) Engineered interfaces of an AAA+ ATPase reveal a new nucleotide-dependent coordination mechanism. J Biol Chem 285:15178–15186
Lyubimov AY, Strycharska M, Berger JM (2011) The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol 21:240–248
Gai D, Zhao R, Li D, Finkielstein CV et al (2004) Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119:47–60
Boyer PD (1989) A perspective of the binding change mechanism for ATP synthesis. FASEB J 3:2164–2178
Stinson BM, Baytshtok V, Schmitz KR, Baker TA et al (2015) Subunit asymmetry and roles of conformational switching in the hexameric AAA+ ring of ClpX. Nat Struct Mol Biol 22:411–416
Danko SJ, Suzuki H (2016) The use of metal fluoride compounds as phosphate analogs for understanding the structural mechanism in P-type ATPases. Methods Mol Biol 1377:195–209
Ogawa H, Cornelius F, Hirata A, Toyoshima C (2015) Sequential substitution of K(+) bound to Na(+), K(+)-ATPase visualized by X-ray crystallography. Nat Commun 6:8004
Wendler P, Saibil HR (2010) Cryo electron microscopy structures of Hsp100 proteins: crowbars in or out? Biochem Cell Biol 88:89–96
Carroni M, Kummer E, Oguchi Y, Wendler P et al (2014) Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. Elife 3:e02481
Fernandez-Millan P, Lazaro M, Cansiz-Arda S, Gerhold JM et al (2015) The hexameric structure of the human mitochondrial replicative helicase Twinkle. Nucleic Acids Res 43:4284–4295
Yeung HO, Forster A, Bebeacua C, Niwa H et al (2014) Inter-ring rotations of AAA ATPase p97 revealed by electron cryomicroscopy. Open Biol 4:130142
Beuron F, Flynn TC, Ma J, Kondo H et al (2003) Motions and negative cooperativity between p97 domains revealed by cryo-electron microscopy and quantised elastic deformational model. J Mol Biol 327:619–629
Banerjee S, Bartesaghi A, Merk A, Rao P et al (2016) 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science 351:871–875
Hanzelmann P, Schindelin H (2016) Structural basis of ATP hydrolysis and intersubunit signaling in the AAA+ ATPase p97. Structure 24:127–139
Schuller JM, Beck F, Lossl P, Heck AJ et al (2016) Nucleotide-dependent conformational changes of the AAA+ ATPase p97 revisited. FEBS Lett 590:595–604
Huang X, Luan B, Wu J, Shi Y (2016) An atomic structure of the human 26S proteasome. Nat Struct Mol Biol 23:778–785
Schweitzer A, Aufderheide A, Rudack T, Beck F et al (2016) Structure of the human 26S proteasome at a resolution of 3.9 A. Proc Natl Acad Sci USA 113:7816–7821
Davies JM, Tsuruta H, May AP, Weis WI (2005) Conformational changes of p97 during nucleotide hydrolysis determined by small-angle X-ray scattering. Structure 13:183–195
Chen B, Doucleff M, Wemmer DE, De Carlo S et al (2007) ATP ground- and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, sigma54. Structure 15:429–440
Roessle M, Manakova E, Laure I, Nawroth T et al (2000) Time-resolved small angle scattering: kinetic and structural data from proteins in solution. J Appl Crystallogr 33:548–551
Kirby NM, Cowieson NP (2014) Time-resolved studies of dynamic biomolecules using small angle X-ray scattering. Curr Opin Struct Biol 28:41–46
Murayama Y, Mukaiyama A, Imai K, Onoue Y et al (2011) Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution. EMBO J 30:68–78
Inobe T, Arai M, Nakao M, Ito K et al (2003) Equilibrium and kinetics of the allosteric transition of GroEL studied by solution X-ray scattering and fluorescence spectroscopy. J Mol Biol 327:183–191
Svergun DI, Stuhrmann HB (1991) New developments in direct shape determination from small-angle scattering. 1. Theory and model-calculations. Acta Crystallogr Sect A 47:736–744
Schneidman-Duhovny D, Kim SJ, Sali A (2012) Integrative structural modeling with small angle X-ray scattering profiles. BMC Struct Biol 12:17
Rambo RP, Tainer JA (2013) Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496:477–481
Valentini E, Kikhney AG, Previtali G, Jeffries CM et al (2015) SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res 43:D357–D363
Huang R, Ripstein ZA, Augustyniak R, Lazniewski M et al (2016) Unfolding the mechanism of the AAA+ unfoldase VAT by a combined cryo-EM, solution NMR study. Proc Natl Acad Sci USA 113:E4190–E4199
Joly N, Zhang N, Buck M (2012) ATPase site architecture is required for self-assembly and remodeling activity of a hexameric AAA+ transcriptional activator. Mol Cell 47:484–490
Eckert T, Link S, Le DT, Sobczak JP et al (2012) Subunit interactions and cooperativity in the microtubule-severing AAA ATPase spastin. J Biol Chem 287:26278–26290
Hoskins JR, Doyle SM, Wickner S (2009) Coupling ATP utilization to protein remodeling by ClpB, a hexameric AAA+ protein. Proc Natl Acad Sci USA 106:22233–22238
Werbeck ND, Schlee S, Reinstein J (2008) Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J Mol Biol 378:178–190
Stotz M, Mueller-Cajar O, Ciniawsky S, Wendler P et al (2011) Structure of green-type Rubisco activase from tobacco. Nat Struct Mol Biol 18:1366–1370
Lundqvist J, Braumann I, Kurowska M, Muller AH et al (2013) Catalytic turnover triggers exchange of subunits of the magnesium chelatase AAA+ motor unit. J Biol Chem 288:24012–24019
Batchelor JD, Doucleff M, Lee CJ, Matsubara K et al (2008) Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation. J Mol Biol 384:1058–1075
De Carlo S, Chen B, Hoover TR, Kondrashkina E et al (2006) The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev 20:1485–1495
Stinson BM, Nager AR, Glynn SE, Schmitz KR et al (2013) Nucleotide binding and conformational switching in the hexameric ring of a AAA+ machine. Cell 153:628–639
Lowe J, Ellonen A, Allen MD, Atkinson C et al (2008) Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol Cell 31:498–509
Crozat E, Meglio A, Allemand JF, Chivers CE et al (2010) Separating speed and ability to displace roadblocks during DNA translocation by FtsK. EMBO J 29:1423–1433
Chen Z, Yang H, Pavletich NP (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453:489–494
Glynn SE, Nager AR, Baker TA, Sauer RT (2012) Dynamic and static components power unfolding in topologically closed rings of a AAA+ proteolytic machine. Nat Struct Mol Biol 19:616–622
Biter AB, Lee S, Sung N, Tsai FT (2012) Structural basis for intersubunit signaling in a protein disaggregating machine. Proc Natl Acad Sci USA 109:12515–12520
Zakeri B, Fierer JO, Celik E, Chittock EC et al (2012) Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci USA 109:E690–E697
Popp MW, Antos JM, Grotenbreg GM, Spooner E et al (2007) Sortagging: a versatile method for protein labeling. Nat Chem Biol 3:707–708
Liu S, Chistol G, Bustamante C (2014) Mechanical operation and intersubunit coordination of ring-shaped molecular motors: insights from single-molecule studies. Biophys J 106:1844–1858
Besprozvannaya M, Burton BM (2014) Do the same traffic rules apply? Directional chromosome segregation by SpoIIIE and FtsK. Mol Microbiol 93:599–608
Olivares AO, Baker TA, Sauer RT (2016) Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol 14:33–44
Belyy V, Yildiz A (2014) Processive cytoskeletal motors studied with single-molecule fluorescence techniques. FEBS Lett 588:3520–3525
Chemla YR, Smith DE (2012) Single-molecule studies of viral DNA packaging. Adv Exp Med Biol 726:549–584
Fili N (2014) Single-molecule and single-particle imaging of molecular motors in vitro and in vivo. EXS 105:131–159
Taraska JW, Puljung MC, Olivier NB, Flynn GE et al (2009) Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat Methods 6:532–537
Kim YC, Snoberger A, Schupp J, Smith DM (2015) ATP binding to neighbouring subunits and intersubunit allosteric coupling underlie proteasomal ATPase function. Nat Commun 6:8520
Aker J, Hesselink R, Engel R, Karlova R et al (2007) In vivo hexamerization and characterization of the Arabidopsis AAA ATPase CDC48A complex using Forster resonance energy transfer-fluorescence lifetime imaging microscopy and fluorescence correlation spectroscopy. Plant Physiol 145:339–350
Zhang N, Gordiyenko Y, Joly N, Lawton E et al (2014) Subunit dynamics and nucleotide-dependent asymmetry of an AAA(+) transcription complex. J Mol Biol 426:71–83
Dyachenko A, Gruber R, Shimon L, Horovitz A et al (2013) Allosteric mechanisms can be distinguished using structural mass spectrometry. Proc Natl Acad Sci USA 110:7235–7239
Rand KD, Zehl M, Jorgensen TJ (2014) Measuring the hydrogen/deuterium exchange of proteins at high spatial resolution by mass spectrometry: overcoming gas-phase hydrogen/deuterium scrambling. Acc Chem Res 47:3018–3027
Konermann L, Pan J, Liu YH (2011) Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev 40:1224–1234
Zhang Q, Chen J, Kuwajima K, Zhang HM et al (2013) Nucleotide-induced conformational changes of tetradecameric GroEL mapped by H/D exchange monitored by FT-ICR mass spectrometry. Sci Rep 3:1247
Chodavarapu S, Jones AD, Feig M, Kaguni JM (2016) DnaC traps DnaB as an open ring and remodels the domain that binds primase. Nucleic Acids Res 44:210–220
Snijder J, Burnley RJ, Wiegard A, Melquiond AS et al (2014) Insight into cyanobacterial circadian timing from structural details of the KaiB-KaiC interaction. Proc Natl Acad Sci USA 111:1379–1384
Ando T (2013) Molecular machines directly observed by high-speed atomic force microscopy. FEBS Lett 587:997–1007
Ando T (2013) High-speed atomic force microscopy. Microscopy (Oxf) 62:81–93
Eghiaian F, Rico F, Colom A, Casuso I et al (2014) High-speed atomic force microscopy: imaging and force spectroscopy. FEBS Lett 588:3631–3638
Noi K, Yamamoto D, Nishikori S, Arita-Morioka K et al (2013) High-speed atomic force microscopic observation of ATP-dependent rotation of the AAA+ chaperone p97. Structure 21:1992–2002
Uchihashi T, Iino R, Ando T, Noji H (2011) High-speed atomic force microscopy reveals rotary catalysis of rotorless F(1)-ATPase. Science 333:755–758
Bujalowski W, Klonowska MM, Jezewska MJ (1994) Oligomeric structure of Escherichia coli primary replicative helicase DnaB protein. J Biol Chem 269:31350–31358
Hurley JH, Yang B (2014) Making sense of Vps4. J Mol Biol 426:503–506
Wang J, Song JJ, Franklin MC, Kamtekar S et al (2001) Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure 9:177–184
Lo YH, Tsai KL, Sun YJ, Chen WT et al (2009) The crystal structure of a replicative hexameric helicase DnaC and its complex with single-stranded DNA. Nucleic Acids Res 37:804–814
Acknowledgments
I would like to thank Drs. Marina Besprozvannaya, Lia Cardarelli, Baoyu Chen, Prashanti Iyer, Ryan Tsoi, and my anonymous reviewers for their helpful comments and suggestions on the manuscript. I apologize for not including a significant number of relevant and detailed studies from my colleagues in the field on account of limited space.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sysoeva, T.A. Assessing heterogeneity in oligomeric AAA+ machines. Cell. Mol. Life Sci. 74, 1001–1018 (2017). https://doi.org/10.1007/s00018-016-2374-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2374-z