Abstract
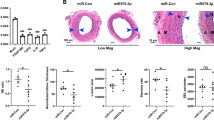
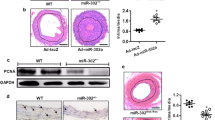
MicroRNAs (miRNAs) coordinate vascular repair by regulating injury-induced gene expression in vascular smooth muscle cells (SMCs) and promote the transition of SMCs from a contractile to a proliferating phenotype. However, the effect of miRNA expression in SMCs on neointima formation is unclear. Therefore, we studied the role of miRNA biogenesis by Dicer in SMCs in vascular repair. Following wire-induced injury to carotid arteries of Apolipoprotein E knockout (Apoe −/−) mice, miRNA microarray analysis revealed that the most significantly regulated miRNAs, such as miR-222 and miR-21-3p, were upregulated. Conditional deletion of Dicer in SMCs increased neointima formation by reducing SMC proliferation in Apoe −/− mice, and decreased mainly the expression of miRNAs, such as miR-147 and miR-100, which were not upregulated following vascular injury. SMC-specific deletion of Dicer promoted growth factor and inflammatory signaling and regulated a miRNA–target interaction network in injured arteries that was enriched in anti-proliferative miRNAs. The most connected miRNA in this network was miR-27a-3p [e.g., with Rho guanine nucleotide exchange factor 26 (ARHGEF26)], which was expressed in medial and neointimal SMCs in a Dicer-dependent manner. In vitro, miR-27a-3p suppresses ARHGEF26 expression and inhibits SMC proliferation by interacting with a conserved binding site in the 3′ untranslated region of ARHGEF26 mRNA. We propose that Dicer expression in SMCs plays an essential role in vascular repair by generating anti-proliferative miRNAs, such as miR-27a-3p, to prevent vessel stenosis due to exaggerated neointima formation.







Similar content being viewed by others
Abbreviations
- Apoe :
-
Apolipoprotein E
- ARHGEF26 :
-
Rho guanine nucleotide exchange factor 26
- CHST1 :
-
Carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
- DAPI:
-
4’,6-Diamidino-2-phenylindole
- DLL4 :
-
Delta-like 4
- DMEM:
-
Dulbecco′s modified eagle medium
- EC:
-
Endothelial cell
- EGF:
-
Epidermal growth factor
- HASMC:
-
Human aortic smooth muscle cell
- HEK293:
-
Human embryonic kidney 293 cell
- HFD:
-
High-fat diet
- IGFBP3 :
-
Insulin-like growth-factor-binding protein 3
- IL-1β:
-
Interleukin-1β
- KO:
-
Knockout
- LNA:
-
Locked nucleic acid
- microRNA:
-
miRNA, miR
- MYH11 :
-
Myosin, heavy chain 11, smooth muscle
- NF-κB:
-
Nuclear factor of kappa light polypeptide gene enhancer in B-cells
- OIT3 :
-
Oncoprotein-induced transcript 3
- PDGF:
-
Platelet-derived growth factor
- miRISC:
-
miRNA-induced-silencing complex
- SH3BGRL2 :
-
SH3-domain-binding glutamate-rich protein-like 2
- SMA:
-
Smooth muscle actin
- SMC:
-
Smooth muscle cell
- TAGLN :
-
Transgelin
- TNFα:
-
Tumor necrosis factor α
- TNRC6A:
-
Argonaute and trinucleotide repeat containing 6A
- TSB:
-
Target site blocker
- UTR:
-
Untranslated region
- WT:
-
Wild type
References
Owens GK, Kumar MS, Wamhoff BR (2004) Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 84:767–801
Alfonso F, Byrne RA, Rivero F, Kastrati A (2014) Current treatment of in-stent restenosis. J Am Coll Cardiol 63:2659–2673. doi:10.1016/j.jacc.2014.02.545
Liu B, Fisher M, Groves P (2002) Down-regulation of the ERK1 and ERK2 mitogen-activated protein kinases using antisense oligonucleotides inhibits intimal hyperplasia in a porcine model of coronary balloon angioplasty. Cardiovasc Res 54:640–648
Stabile E, Zhou YF, Saji M, Castagna M, Shou M, Kinnaird TD, Baffour R, Ringel MD, Epstein SE, Fuchs S (2003) Akt controls vascular smooth muscle cell proliferation in vitro and in vivo by delaying G1/S exit. Circ Res 93:1059–1065. doi:10.1161/01.res.0000105086.31909.1b
Peppel K, Zhang L, Orman ES, Hagen PO, Amalfitano A, Brian L, Freedman NJ (2005) Activation of vascular smooth muscle cells by TNF and PDGF: overlapping and complementary signal transduction mechanisms. Cardiovasc Res 65:674–682. doi:10.1016/j.cardiores.2004.10.031
Yoshida T, Yamashita M, Horimai C, Hayashi M (2013) Smooth muscle-selective inhibition of nuclear factor-kappaB attenuates smooth muscle phenotypic switching and neointima formation following vascular injury. J Am Heart Assoc 2:e000230. doi:10.1161/jaha.113.000230
Siciliano V, Garzilli I, Fracassi C, Criscuolo S, Ventre S, di Bernardo D (2013) MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat Commun 4:2364. doi:10.1038/ncomms3364
Ebert MS, Sharp PA (2012) Roles for MicroRNAs in conferring robustness to biological processes. Cell 149:515–524. doi:10.1016/j.cell.2012.04.005
Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524. doi:10.1038/nrm3838
Clark PM, Loher P, Quann K, Brody J, Londin ER, Rigoutsos I (2014) Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types. Sci Rep 4:5947. doi:10.1038/srep05947
Chi SW, Zang JB, Mele A, Darnell RB (2009) Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460:479–486. doi:10.1038/nature08170
Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC (2010) MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol 30:1118–1126. doi:10.1161/atvbaha.109.200873
Albinsson S, Skoura A, Yu J, DiLorenzo A, Fernandez-Hernando C, Offermanns S, Miano JM, Sessa WC (2011) Smooth muscle miRNAs are critical for post-natal regulation of blood pressure and vascular function. PLoS One 6:e18869. doi:10.1371/journal.pone.0018869
Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C (2009) MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res 105:158–166. doi:10.1161/circresaha.109.197517
Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C (2007) MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res 100:1579–1588
Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN (2009) MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev 23:2166–2178. doi:10.1101/gad.1842409
McDonald RA, Hata A, MacLean MR, Morrell NW, Baker AH (2012) MicroRNA and vascular remodelling in acute vascular injury and pulmonary vascular remodelling. Cardiovasc Res 93:594–604. doi:10.1093/cvr/cvr299
Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ (2005) The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA 102:10898–10903
Wirth A, Benyo Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, Orsy P, Horvath B, Maser-Gluth C, Greiner E, Lemmer B, Schutz G, Gutkind JS, Offermanns S (2008) G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med 14:64–68
Schober A, Knarren S, Lietz M, Lin EA, Weber C (2003) Crucial role of stromal cell-derived factor-1alpha in neointima formation after vascular injury in apolipoprotein E-deficient mice. Circulation 108:2491–2497. doi:10.1161/01.CIR.0000099508.76665.9A
Bisognin A, Sales G, Coppe A, Bortoluzzi S, Romualdi C (2012) MAGIA2: from miRNA and genes expression data integrative analysis to microRNA-transcription factor mixed regulatory circuits (2012 update). Nucleic Acids Res. doi:10.1093/nar/gks460
Nuovo GJ, Elton TS, Nana-Sinkam P, Volinia S, Croce CM, Schmittgen TD (2009) A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc 4:107–115
Cambronne XA, Shen R, Auer PL, Goodman RH (2012) Capturing microRNA targets using an RNA-induced silencing complex (RISC)-trap approach. Proc Natl Acad Sci USA 109:20473–20478. doi:10.1073/pnas.1218887109
Nelson M, Mcclelland M (1992) Use of DNA methyltransferase endonuclease enzyme combinations for megabase mapping of chromosomes. Method Enzymol 216:279–303
Hartmann P, Zhou Z, Natarelli L, Wei Y, Nazari-Jahantigh M, Zhu M, Grommes J, Steffens S, Weber C, Schober A (2016) Endothelial Dicer promotes atherosclerosis and vascular inflammation by miRNA-103-mediated suppression of KLF4. Nat Commun 7:10521. doi:10.1038/ncomms10521
Schober A, Nazari-Jahantigh M, Weber C (2015) MicroRNA-mediated mechanisms of the cellular stress response in atherosclerosis. Nat Rev Cardiol 12:361–374. doi:10.1038/nrcardio.2015.38
Cook CL, Weiser MC, Schwartz PE, Jones CL, Majack RA (1994) Developmentally timed expression of an embryonic growth phenotype in vascular smooth muscle cells. Circ Res 74:189–196
Chen CN, Li YS, Yeh YT, Lee PL, Usami S, Chien S, Chiu JJ (2006) Synergistic roles of platelet-derived growth factor-BB and interleukin-1beta in phenotypic modulation of human aortic smooth muscle cells. Proc Natl Acad Sci USA 103:2665–2670. doi:10.1073/pnas.0510973103
Leeper NJ, Raiesdana A, Kojima Y, Chun HJ, Azuma J, Maegdefessel L, Kundu RK, Quertermous T, Tsao PS, Spin JM (2011) MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol 226:1035–1043. doi:10.1002/jcp.22422
Fiedler J, Stohr A, Gupta SK, Hartmann D, Holzmann A, Just A, Hansen A, Hilfiker-Kleiner D, Eschenhagen T, Thum T (2014) Functional microRNA library screening identifies the hypoxamir miR-24 as a potent regulator of smooth muscle cell proliferation and vascularization. Antioxid Redox Signal 21:1167–1176. doi:10.1089/ars.2013.5418
Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D (2009) miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460:705–710. doi:10.1038/nature08195
Choe N, Kwon JS, Kim JR, Eom GH, Kim Y, Nam KI, Ahn Y, Kee HJ, Kook H (2013) The microRNA miR-132 targets Lrrfip1 to block vascular smooth muscle cell proliferation and neointimal hyperplasia. Atherosclerosis 229:348–355. doi:10.1016/j.atherosclerosis.2013.05.009
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23:4051–4060. doi:10.1038/sj.emboj.7600385
Hernandez-Torres F, Aranega AE, Franco D (2014) Identification of regulatory elements directing miR-23a-miR-27a-miR-24-2 transcriptional regulation in response to muscle hypertrophic stimuli. Biochim Biophys Acta 1839:885–897. doi:10.1016/j.bbagrm.2014.07.009
Ellerbroek SM, Wennerberg K, Arthur WT, Dunty JM, Bowman DR, DeMali KA, Der C, Burridge K (2004) SGEF, a RhoG guanine nucleotide exchange factor that stimulates macropinocytosis. Mol Biol Cell 15:3309–3319. doi:10.1091/mbc.E04-02-0146
Wang H, Wu R, Yu L, Wu F, Li S, Zhao Y, Li H, Luo G, Wang J, Zhou J (2012) SGEF is overexpressed in prostate cancer and contributes to prostate cancer progression. Oncol Rep 28:1468–1474. doi:10.3892/or.2012.1917
Wang H, Li S, Li H, Wang P, Huang F, Zhao Y, Yu L, Luo G, Zhang X, Wang J, Zhou J (2014) Grb2 interacts with SGEF and antagonizes the ability of SGEF to enhance EGF-induced ERK1/2 activation. Mol Cell Biochem 389:239–247. doi:10.1007/s11010-013-1945-7
Tang RH, Zheng XL, Callis TE, Stansfield WE, He J, Baldwin AS, Wang DZ, Selzman CH (2008) Myocardin inhibits cellular proliferation by inhibiting NF-kappaB(p65)-dependent cell cycle progression. Proc Natl Acad Sci USA 105:3362–3367. doi:10.1073/pnas.0705842105
Acknowledgments
This work has been funded by the German Research Foundation (DFG) as part of the Collaborative Research Center 1123 (B04) and by the German Center for Cardiovascular Research (MHA VD1.2). The authors declare no competing financial interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
F. Zahedi and M. Nazari-Jahantigh are co-first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zahedi, F., Nazari-Jahantigh, M., Zhou, Z. et al. Dicer generates a regulatory microRNA network in smooth muscle cells that limits neointima formation during vascular repair. Cell. Mol. Life Sci. 74, 359–372 (2017). https://doi.org/10.1007/s00018-016-2349-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2349-0