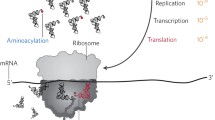
Abstract
Regulation of protein synthesis contributes to maintenance of homeostasis and adaptation to environmental changes. mRNA translation is controlled at various levels including initiation, elongation and termination, through post-transcriptional/translational modifications of components of the protein synthesis machinery. Recently, protein and RNA hydroxylation have emerged as important enzymatic modifications of tRNAs, elongation and termination factors, as well as ribosomal proteins. These modifications enable a correct STOP codon recognition, ensuring translational fidelity. Recent studies are starting to show that STOP codon read-through is related to the ability of the cell to cope with different types of stress, such as oxidative and chemical insults, while correlations between defects in hydroxylation of protein synthesis components and STOP codon read-through are beginning to emerge. In this review we will discuss our current knowledge of protein synthesis regulation through hydroxylation of components of the translation machinery, with special focus on STOP codon recognition. We speculate on the possibility that programmed STOP codon read-through, modulated by hydroxylation of components of the protein synthesis machinery, is part of a concerted cellular response to stress.



Similar content being viewed by others
References
Aoyagi Y et al (1988) Energy cost of whole-body protein synthesis measured in vivo in chicks. Comp Biochem Physiol A Comp Physiol 91(4):765–768
Bock FJ, Todorova TT, Chang P (2015) RNA regulation by poly(ADP-Ribose) polymerases. Mol Cell 58(6):959–969
Kang SA et al (2013) mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341(6144):1236566
Carroll AJ et al (2008) Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol Cell Proteomics 7(2):347–369
Kearse MG, Chen AS, Ware VC (2011) Expression of ribosomal protein L22e family members in Drosophila melanogaster: rpL22-like is differentially expressed and alternatively spliced. Nucleic Acids Res 39(7):2701–2716
Lopes AM et al (2010) The human RPS4 paralogue on Yq11.223 encodes a structurally conserved ribosomal protein and is preferentially expressed during spermatogenesis. BMC Mol Biol 11:33
Hinnebusch AG (2014) The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83:779–812
Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11(2):113–127
Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136(4):731–745
Rodnina MV, Beringer M, Wintermeyer W (2007) How ribosomes make peptide bonds. Trends Biochem Sci 32(1):20–26
Nurenberg E, Tampe R (2013) Tying up loose ends: ribosome recycling in eukaryotes and archaea. Trends Biochem Sci 38(2):64–74
Young DJ et al (2015) Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo. Cell 162(4):872–884
Passmore LA et al (2007) The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26(1):41–50
Kunze G et al (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 16(12):3496–3507
Dzialo MC et al (2014) Translational roles of elongation factor 2 protein lysine methylation. J Biol Chem 289(44):30511–30524
Odintsova TI et al (2003) Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing. J Protein Chem 22(3):249–258
Arnold RJ, Reilly JP (2002) Analysis of methylation and acetylation in E. coli ribosomal proteins. Methods Mol Biol 194:205–210
Lee SW et al (2002) Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc Natl Acad Sci USA 99(9):5942–5947
Cammarano P et al (1972) Characterization of unfolded and compact ribosomal subunits from plants and their relationship to those of lower and higher animals: evidence for physicochemical heterogeneity among eucaryotic ribosomes. Biochim Biophys Acta 281(4):571–596
Louie DF et al (1996) Mass spectrometric analysis of 40S ribosomal proteins from Rat-1 fibroblasts. J Biol Chem 271(45):28189–28198
Vladimirov SN et al (1996) Characterization of the human small-ribosomal-subunit proteins by N-terminal and internal sequencing, and mass spectrometry. Eur J Biochem 239(1):144–149
Ban N et al (2014) A new system for naming ribosomal proteins. Curr Opin Struct Biol 24:165–169
Ruvinsky I et al (2005) Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 19(18):2199–2211
Fu Y et al (2010) The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew Chem Int Ed Engl 49(47):8885–8888
Kato M et al (2011) Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification. Nucleic Acids Res 39(4):1576–1585
Scotti JS et al (2014) Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation. Proc Natl Acad Sci USA 111(37):13331–13336
Feng T et al (2014) Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol Cell 53(4):645–654
Ge W et al (2012) Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol 8(12):960–962
van Staalduinen LM, Novakowski SK, Jia Z (2014) Structure and functional analysis of YcfD, a novel 2-oxoglutarate/Fe(2)(+)-dependent oxygenase involved in translational regulation in Escherichia coli. J Mol Biol 426(9):1898–1910
Loenarz C, Schofield CJ (2011) Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci 36(1):7–18
Markolovic S, Wilkins SE, Schofield CJ (2015) Protein hydroxylation catalyzed by 2-oxoglutarate-dependent oxygenases. J Biol Chem 290(34):20712–20722
Bugg TD (2001) Oxygenases: mechanisms and structural motifs for O(2) activation. Curr Opin Chem Biol 5(5):550–555
Mittal N et al (2013) Unique posttranslational modifications in eukaryotic translation factors and their roles in protozoan parasite viability and pathogenesis. Mol Biochem Parasitol 187(1):21–31
Dever TE, Green R (2012) The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 4(7):a013706
Gupta N et al (2007) Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res 17(9):1362–1377
Blaha G, Stanley RE, Steitz TA (2009) Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science 325(5943):966–970
Pavlov MY et al (2009) Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci USA 106(1):50–54
Ude S et al (2013) Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339(6115):82–85
Doerfel LK et al (2013) EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339(6115):85–88
Roy H et al (2011) The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-beta-lysine. Nat Chem Biol 7(10):667–669
Peil L et al (2012) Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat Chem Biol 8(8):695–697
Navarre WW et al (2010) PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell 39(2):209–221
Bullwinkle TJ et al (2013) (R)-Beta-lysine-modified elongation factor P functions in translation elongation. J Biol Chem 288(6):4416–4423
Kemper WM, Berry KW, Merrick WC (1976) Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Balpha and M2Bbeta. J Biol Chem 251(18):5551–5557
Gregio AP et al (2009) eIF5A has a function in the elongation step of translation in yeast. Biochem Biophys Res Commun 380(4):785–790
Kang HA, Hershey JW (1994) Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem 269(6):3934–3940
Saini P et al (2009) Hypusine-containing protein eIF5A promotes translation elongation. Nature 459(7243):118–121
Dever TE, Gutierrez E, Shin BS (2014) The hypusine-containing translation factor eIF5A. Crit Rev Biochem Mol Biol 49(5):413–425
Gutierrez E et al (2013) eIF5A promotes translation of polyproline motifs. Mol Cell 51(1):35–45
Park MH, Joe YA, Kang KR (1998) Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J Biol Chem 273(3):1677–1683
Schrader R et al (2006) Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J Biol Chem 281(46):35336–35346
Park JH et al (2006) Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc Natl Acad Sci USA 103(1):51–56
Patel PH et al (2009) The Drosophila deoxyhypusine hydroxylase homologue nero and its target eIF5A are required for cell growth and the regulation of autophagy. J Cell Biol 185(7):1181–1194
Sievert H et al (2014) A novel mouse model for inhibition of DOHH-mediated hypusine modification reveals a crucial function in embryonic development, proliferation and oncogenic transformation. Dis Model Mech 7(8):963–976
Taylor D et al (2012) Cryo-EM structure of the mammalian eukaryotic release factor eRF1-eRF3-associated termination complex. Proc Natl Acad Sci USA 109(45):18413–18418
Carlson BA et al (1999) Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology 255(1):2–8
Varani G, McClain WH (2000) The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep 1(1):18–23
Gorgoni B et al (2014) Controlling translation elongation efficiency: tRNA regulation of ribosome flux on the mRNA. Biochem Soc Trans 42(1):160–165
Ogle JM et al (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292(5518):897–902
Sharma D et al (2007) Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J Mol Biol 374(4):1065–1076
van den Born E et al (2011) ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun 2:172
Kawakami M et al (1988) Chemical structure of a new modified nucleoside located in the anticodon of Bombyx mori glycine tRNA2. J Biochem 104(1):108–111
Kawakami M et al (1980) Abnormal codon recognition of glycyl-tRNA from the posterior silk glands of Bombyx mori. J Biochem 88(4):1151–1157
Konevega AL et al (2004) Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA 10(1):90–101
Noma A et al (2010) Expanding role of the jumonji C domain as an RNA hydroxylase. J Biol Chem 285(45):34503–34507
Chowdhury R et al (2014) Ribosomal oxygenases are structurally conserved from prokaryotes to humans. Nature 510(7505):422–426
Suzuki C et al (2007) Identification of Myc-associated protein with JmjC domain as a novel therapeutic target oncogene for lung cancer. Mol Cancer Ther 6(2):542–551
Teye K et al (2004) Increased expression of a Myc target gene Mina53 in human colon cancer. Am J Pathol 164(1):205–216
Wang C et al (2015) Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8. Acta Crystallogr D Biol Crystallogr 71(Pt 9):1955–1964
Yanshina DD et al (2015) Hydroxylated histidine of human ribosomal protein uL2 is involved in maintaining the local structure of 28S rRNA in the ribosomal peptidyl transferase center. FEBS J 282(8):1554–1566
Singleton RS et al (2014) OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc Natl Acad Sci USA 111(11):4031–4036
Loenarz C et al (2014) Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc Natl Acad Sci USA 111(11):4019–4024
Katz MJ et al (2014) Sudestada1, a Drosophila ribosomal prolyl-hydroxylase required for mRNA translation, cell homeostasis, and organ growth. Proc Natl Acad Sci USA 111(11):4025–4030
Keeling KM et al (2006) Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol 26(14):5237–5248
Torabi N, Kruglyak L (2012) Genetic basis of hidden phenotypic variation revealed by increased translational readthrough in yeast. PLoS Genet 8(3):e1002546
Namy O, Duchateau-Nguyen G, Rousset JP (2002) Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol Microbiol 43(3):641–652
Freitag J, Ast J, Bolker M (2012) Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485(7399):522–525
Touriol C et al (2003) Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell 95(3–4):169–178
Casey JL, Gerin JL (1995) Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J Virol 69(12):7593–7600
Jacks T, Varmus HE (1985) Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science 230(4731):1237–1242
Weiner AM, Weber K (1971) Natural read-through at the UGA termination signal of Q-beta coat protein cistron. Nat New Biol 234(50):206–209
Geller AI, Rich A (1980) A UGA termination suppression tRNATrp active in rabbit reticulocytes. Nature 283(5742):41–46
Yamaguchi Y et al (2012) L-MPZ, a novel isoform of myelin P0, is produced by stop codon readthrough. J Biol Chem 287(21):17765–17776
Beier H, Grimm M (2001) Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res 29(23):4767–4782
Stiebler AC et al (2014) Ribosomal readthrough at a short UGA stop codon context triggers dual localization of metabolic enzymes in Fungi and animals. PLoS Genet 10(10):e1004685
Jungreis I et al (2011) Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res 21(12):2096–2113
Brown CM et al (1993) The translational termination signal database. Nucleic Acids Res 21(13):3119–3123
Tate WP et al (1995) Translational termination efficiency in both bacteria and mammals is regulated by the base following the stop codon. Biochem Cell Biol 73(11–12):1095–1103
Namy O, Hatin I, Rousset JP (2001) Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep 2(9):787–793
Berry MJ et al (1991) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353(6341):273–276
Kossinova O et al (2013) A novel insight into the mechanism of mammalian selenoprotein synthesis. RNA 19(8):1147–1158
Jalajakumari MB et al (1989) Genes for biosynthesis and assembly of CS3 pili of CFA/II enterotoxigenic Escherichia coli: novel regulation of pilus production by bypassing an amber codon. Mol Microbiol 3(12):1685–1695
Schueren F et al (2014) Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. Elife 3:e03640
Hofstetter H, Monstein HJ, Weissmann C (1974) The readthrough protein A1 is essential for the formation of viable Q beta particles. Biochim Biophys Acta 374(2):238–251
Pelham HR (1978) Leaky UAG termination codon in tobacco mosaic virus RNA. Nature 272(5652):469–471
Philipson L et al (1978) Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell 13(1):189–199
Yoshinaka Y et al (1985) Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci USA 82(6):1618–1622
Boylan M, Coleman DC, Smyth CJ (1987) Molecular cloning and characterization of the genetic determinant encoding CS3 fimbriae of enterotoxigenic Escherichia coli. Microb Pathog 2(3):195–209
Lin MF et al (2007) Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res 17(12):1823–1836
Dunn JG et al (2013) Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife 2:e01179
Xue F, Cooley L (1993) kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72(5):681–693
Steneberg P et al (1998) Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the drosophila trachea. Genes Dev 12(7):956–967
Bergstrom DE et al (1995) Regulatory autonomy and molecular characterization of the Drosophila out at first gene. Genetics 139(3):1331–1346
Klagges BR et al (1996) Invertebrate synapsins: a single gene codes for several isoforms in Drosophila. J Neurosci 16(10):3154–3165
Samuels ME, Schedl P, Cline TW (1991) The complex set of late transcripts from the Drosophila sex determination gene sex-lethal encodes multiple related polypeptides. Mol Cell Biol 11(7):3584–3602
Samson ML, Lisbin MJ, White K (1995) Two distinct temperature-sensitive alleles at the elav locus of Drosophila are suppressed nonsense mutations of the same tryptophan codon. Genetics 141(3):1101–1111
Chao AT et al (2003) Mutations in eukaryotic release factors 1 and 3 act as general nonsense suppressors in Drosophila. Genetics 165(2):601–612
Chittum HS et al (1998) Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry 37(31):10866–10870
Eswarappa SM et al (2014) Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell 157(7):1605–1618
Carmeliet P et al (1996) Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 380(6573):435–439
Demirci H et al (2013) The central role of protein S12 in organizing the structure of the decoding site of the ribosome. RNA 19(12):1791–1801
Ogle JM et al (2002) Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111(5):721–732
Demirci H et al (2013) A structural basis for streptomycin-induced misreading of the genetic code. Nat Commun 4:1355
O’Connor M et al (1995) Genetic probes of ribosomal RNA function. Biochem Cell Biol 73(11–12):859–868
Funatsu G, Wittmann HG (1972) Ribosomal proteins. 33. Location of amino-acid replacements in protein S12 isolated from Escherichia coli mutants resistant to streptomycin. J Mol Biol 68(3):547–550
Alksne LE et al (1993) An accuracy center in the ribosome conserved over 2 billion years. Proc Natl Acad Sci USA 90(20):9538–9541
Gorini L, Kataja E (1964) Streptomycin-induced oversuppression in E. coli. Proc Natl Acad Sci USA 51:995–1001
Chandramouli P et al (2008) Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure 16(4):535–548
Bonetti B et al (1995) The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol 251(3):334–345
Selmer M et al (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313(5795):1935–1942
Noeske J et al (2015) High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol 22(4):336–341. doi:10.1038/nsmb.2994
Nakamura Y, Ito K (2011) tRNA mimicry in translation termination and beyond. Wiley Interdiscip Rev RNA 2(5):647–668
Chavatte L et al (2002) The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J 21(19):5302–5311
Bulygin KN et al (2010) Three distinct peptides from the N domain of translation termination factor eRF1 surround stop codon in the ribosome. RNA 16(10):1902–1914
Brown A et al (2015) Structural basis for stop codon recognition in eukaryotes. Nature 524(7566):493–496
Laplante M, Sabatini DM (2012) mTOR signaling in growth control and disease. Cell 149(2):274–293
Vannuvel K et al (2013) Functional and morphological impact of ER stress on mitochondria. J Cell Physiol 228(9):1802–1818
Pimentel J, Boccaccio GL (2014) Translation and silencing in RNA granules: a tale of sand grains. Front Mol Neurosci 7:68
Thomas MG et al (2011) RNA granules: the good, the bad and the ugly. Cell Signal 23(2):324–334
Bar-Peled L, Sabatini DM (2014) Regulation of mTORC1 by amino acids. Trends Cell Biol 24(7):400–406
Meyuhas O (2008) Physiological roles of ribosomal protein S6: one of its kind. Int Rev Cell Mol Biol 268:1–37
Browne GJ, Proud CG (2004) A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol Cell Biol 24(7):2986–2997
Keegan LP, Gallo A, O’Connell MA (2001) The many roles of an RNA editor. Nat Rev Genet 2(11):869–878
Yamasaki S, Anderson P (2008) Reprogramming mRNA translation during stress. Curr Opin Cell Biol 20(2):222–226
Gerashchenko MV, Lobanov AV, Gladyshev VN (2012) Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci USA 109(43):17394–17399
Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147(4):789–802
Oren YS et al (2014) The unfolded protein response affects readthrough of premature termination codons. EMBO Mol Med 6(5):685–701
Sideri TC et al (2010) Ribosome-associated peroxiredoxins suppress oxidative stress-induced de novo formation of the [PSI+] prion in yeast. Proc Natl Acad Sci USA 107(14):6394–6399
Eaglestone SS, Cox BS, Tuite MF (1999) Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J 18(7):1974–1981
True HL, Berlin I, Lindquist SL (2004) Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431(7005):184–187
Ivan M et al (2001) HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292(5516):464–468
Jaakkola P et al (2001) Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292(5516):468–472
Lando D et al (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 16(12):1466–1471
Bruick RK, McKnight SL (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294(5545):1337–1340
Ratcliffe PJ (2013) Oxygen sensing and hypoxia signalling pathways in animals: the implications of physiology for cancer. J Physiol 591(Pt 8):2027–2042
Epstein AC et al (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107(1):43–54
Karijolich J, Yu YT (2014) Therapeutic suppression of premature termination codons: mechanisms and clinical considerations (review). Int J Mol Med 34(2):355–362
Keeling KM, Bedwell DM (2011) Suppression of nonsense mutations as a therapeutic approach to treat genetic diseases. Wiley Interdiscip Rev RNA 2(6):837–852
Acknowledgments
We wish to thank all members of the Wappner’s lab for discussions and to Peter Ratcliffe for his continuous support. This work was funded by ANPCyT Grants PICT 2014 No. 0649 and PICT 2012 No. 0214. PW is a career investigator of CONICET.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Katz, M.J., Gándara, L., De Lella Ezcurra, A.L. et al. Hydroxylation and translational adaptation to stress: some answers lie beyond the STOP codon. Cell. Mol. Life Sci. 73, 1881–1893 (2016). https://doi.org/10.1007/s00018-016-2160-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2160-y