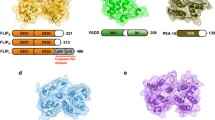
Abstract
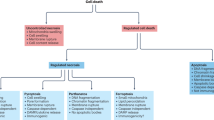
Programmed cell death plays a central role in the regulation of homeostasis and development of multicellular organisms. Deregulation of programmed cell death is connected to a number of disorders, including cancer and autoimmune diseases. Initiation of cell death occurs in the multiprotein complexes or high molecular weight platforms. Composition, structure, and molecular interactions within these platforms influence the cellular decision toward life or death and, therefore, define the induction of a particular cell death program. Here, we discuss in detail the key cell-death complexes—including DISC, complex II, and TNFRI complex I/II, and the necrosome, RIPoptosome, apoptosome, and PIDDosome—that control apoptosis or necroptosis pathways as well as their regulation. The possibility of their pharmacological targeting leading to the development of new strategies of interference with cell death programs via control of the high molecular weight platforms will be discussed.

Similar content being viewed by others
References
Vaux DL, Korsmeyer SJ (1999) Cell death in development. Cell 96:245–254
Kroemer G, Galluzzi L, Vandenabeele P et al (2009) Classification of cell death: recommendations of the nomenclature committee on cell death 2009. Cell Death Differ 16:3–11
Krammer PH, Arnold R, Lavrik IN (2007) Life and death in peripheral T cells. Nat Rev Immunol 7:532–542
Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G (2010) Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 11:700–714
Galluzzi L, Kepp O, Krautwald S, Kroemer G, Linkermann A (2014) Molecular mechanisms of regulated necrosis. Semin Cell Dev Biol 35:24–32
Miura M, Zhu H, Rotello R, Hartwieg EA, Yuan J (1993) Induction of apoptosis in fibroblasts by IL-1 beta-converting enzyme, a mammalian homolog of the C. elegans cell death gene ced-3. Cell 75:653–660
Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR (1993) The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75:641–652
Kuranaga E (2012) Beyond apoptosis: caspase regulatory mechanisms and functions in vivo. Genes Cells 17:83–97
Ciraci C, Janczy JR, Sutterwala FS, Cassel SL (2012) Control of innate and adaptive immunity by the inflammasome. Microbes Infect 14:1263–1270
Degterev A, Boyce M, Yuan J (2003) A decade of caspases. Oncogene 22:8543–8567
Lamkanfi M, Dixit VM (2012) Inflammasomes and their roles in health and disease. Annu Rev Cell Dev Biol 28:137–161
Mittl PR, Di Marco S, Krebs JF et al (1997) Structure of recombinant human CPP32 in complex with the tetrapeptide acetyl-Asp-Val-Ala-Asp fluoromethyl ketone. J Biol Chem 272:6539–6547
Wei Y, Fox T, Chambers SP et al (2000) The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem Biol 7:423–432
Riedl SJ, Renatus M, Schwarzenbacher R et al (2001) Structural basis for the inhibition of caspase-3 by XIAP. Cell 104:791–800
Chai J, Shiozaki E, Srinivasula SM et al (2001) Structural basis of caspase-7 inhibition by XIAP. Cell 104:769–780
Weber CH, Vincenz C (2001) The death domain superfamily: a tale of two interfaces? Trends Biochem Sci 26:475–481
Dickens LS, Powley IR, Hughes MA, MacFarlane M (2012) The, “complexities” of life and death: death receptor signalling platforms. Exp Cell Res 318:1269–1277
Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM (1998) An induced proximity model for caspase-8 activation. J Biol Chem 273:2926–2930
Stennicke HR, Salvesen GS (2000) Caspases: controlling intracellular signals by protease zymogen activation. Biochim Biophys Acta Protein Struct Mol Enzymol 1477:299–306
Muzio M, Chinnaiyan AM, Kischkel FC et al (1996) FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85:817–827
Lavrik IN, Krammer PH (2012) Regulation of CD95/Fas signaling at the DISC. Cell Death Differ 19:36–41
Sessler T, Healy S, Samali A, Szegezdi E (2013) Structural determinants of DISC function: new insights into death receptor-mediated apoptosis signalling. Pharmacol Ther 140:186–199
Schleich K, Warnken U, Fricker N et al (2012) Stoichiometry of the CD95 death-inducing signaling complex: experimental and modeling evidence for a death effector domain chain model. Mol Cell 47:306–319
Dickens LS, Boyd RS, Jukes-Jones R et al (2012) A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Mol Cell 47:291–305
Martin DA, Siegel RM, Zheng L, Lenardo MJ (1998) Membrane oligomerization and cleavage activates the caspase-8 (FLICE/MACHalpha1) death signal. J Biol Chem 273:4345–4349
Chang DW, Xing Z, Capacio VL, Peter ME, Yang X (2003) Interdimer processing mechanism of procaspase-8 activation. EMBO J 22:4132–4142
Hoffmann JC, Pappa A, Krammer PH, Lavrik IN (2009) A new C-terminal cleavage product of procaspase-8, p30, defines an alternative pathway of procaspase-8 activation. Mol Cell Biol 29:4431–4440
Kallenberger SM, Beaudouin J, Claus J et al (2014) Intra- and interdimeric caspase-8 self-cleavage controls strength and timing of CD95-induced apoptosis. Sci Signal 7:ra23
Varfolomeev E, Maecker H, Sharp D et al (2005) Molecular determinants of kinase pathway activation by Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand. J Biol Chem 280:40599–40608
Lavrik IN, Mock T, Golks A, Hoffmann JC, Baumann S, Krammer PH (2008) CD95 stimulation results in the formation of a novel death effector domain protein-containing complex. J Biol Chem 283:26401–26408
Cullen SP, Henry CM, Kearney CJ et al (2013) Fas/CD95-induced chemokines can serve as “Find-Me” signals for apoptotic cells. Mol Cell 49:1034–1048
Geserick P, Hupe M, Moulin M et al (2009) Cellular IAPs inhibit a cryptic CD95-induced cell death by limiting RIP1 kinase recruitment. J Cell Biol 187:1037–1054
Varfolomeev EE, Ashkenazi A (2004) Tumor necrosis factor: an apoptosis JuNKie? Cell 116:491–497
Wajant H, Pfizenmaier K, Scheurich P (2003) Tumor necrosis factor signaling. Cell Death Differ 10:45–65
Micheau O, Tschopp J (2003) Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114:181–190
Haas TL, Emmerich CH, Gerlach B et al (2009) Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and? Is required for TNF-mediated gene induction. Mol Cell 36:831–844
Gerlach B, Cordier SM, Schmukle AC et al (2011) Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471:591–596
Ikeda F, Deribe YL, Skånland SS et al (2011) SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471:637–641
Harper N, Hughes M, MacFarlane M, Cohen GM (2003) Fas-associated death domain protein and caspase-8 are not recruited to the tumor necrosis factor receptor 1 signaling complex during tumor necrosis factor-induced apoptosis. J Biol Chem 278:25534–25541
Oztürk S, Schleich K, Lavrik IN (2012) Cellular FLICE-like inhibitory proteins (c-FLIPs): fine-tuners of life and death decisions. Exp Cell Res 318:1324–1331
Golks A, Brenner D, Fritsch C, Krammer PH, Lavrik IN (2005) c-FLIPR, a new regulator of death receptor-induced apoptosis. J Biol Chem 280:14507–14513
Fricker N, Beaudouin J, Richter P, Eils R, Krammer PH, Lavrik IN (2010) Model-based dissection of CD95 signaling dynamics reveals both a pro- and antiapoptotic role of c-FLIPL. J Cell Biol 190:377–389
Pop C, Oberst A, Drag M et al (2011) FLIP(L) induces caspase 8 activity in the absence of interdomain caspase 8 cleavage and alters substrate specificity. Biochem J 433:447–457
Ofengeim D, Yuan J (2013) Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat Rev Mol Cell Biol 14:727–736
Christofferson DE, Li Y, Yuan J (2014) Control of life-or-death decisions by RIP1 kinase. Annu Rev Physiol 76:129–150
Cho Y, Challa S, Moquin D et al (2009) Phosphorylation-driven assembly of the RIP1–RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137:1112–1123
Li J, McQuade T, Siemer AB et al (2012) The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150:339–350
Moriwaki K, Chan FK-M (2013) RIP3: a molecular switch for necrosis and inflammation. Genes Dev 27:1640–1649
Degterev A, Huang Z, Boyce M et al (2005) Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol 1:112–119
Teng X, Degterev A, Jagtap P et al (2005) Structure-activity relationship study of novel necroptosis inhibitors. Bioorg Med Chem Lett 15:5039–5044
Degterev A, Hitomi J, Germscheid M et al (2008) Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 4:313–321
Choi S, Keys H, Staples RJ, Yuan J, Degterev A, Cuny GD (2012) Optimization of tricyclic Nec-3 necroptosis inhibitors for in vitro liver microsomal stability. Bioorganic Med Chem Lett 22:5685–5688
Sun L, Wang H, Wang Z et al (2012) Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148:213–227
Murphy JM, Czabotar PE, Hildebrand JM et al (2013) The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39:443–453
Dondelinger Y, Declercq W, Montessuit S et al (2014) MLKL compromises plasma membrane integrity upon binding to phosphatidyl inositol phosphates. Cell Rep 7:1–11
Chen X, Li W, Ren J et al (2014) Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res 24:105–121
Wang H, Sun L, Su L et al (2014) Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell 54:133–146
Moujalled DM, Cook WD, Okamoto T et al (2013) TNF can activate RIPK3 and cause programmed necrosis in the absence of RIPK1. Cell Death Dis 4:e465
Feoktistova M, Geserick P, Kellert B et al (2011) CIAPs block ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 43:449–463
Tenev T, Bianchi K, Darding M et al (2011) The ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 43:432–448
Feoktistova M, Geserick P, Panayotova-Dimitrova D, Leverkus M (2012) Pick your poison: the ripoptosome, a cell death platform regulating apoptosis and necroptosis. Cell Cycle 11:460–467
Vanlangenakker N, Bertrand MJM, Bogaert P, Vandenabeele P, Vanden Berghe T (2011) TNF-induced necroptosis in L929 cells is tightly regulated by multiple TNFR1 complex I and II members. Cell Death Dis 2:e230
Remijsen Q, Goossens V, Grootjans S et al (2014) Depletion of RIPK3 or MLKL blocks TNF-driven necroptosis and switches towards a delayed RIPK1 kinase-dependent apoptosis. Cell Death Dis 5:e1004
Danial NN, Korsmeyer SJ (2004) Cell death: critical control points. Cell 116:205–219
Kagan VE, Tyurin VA, Jiang J et al (2005) Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol 1:223–232
Yuan S, Akey CW (2013) Apoptosome structure, assembly, and procaspase activation. Structure 21:501–515
Twiddy D, Brown DG, Adrain C et al (2004) Pro-apoptotic proteins released from the mitochondria regulate the protein composition and caspase-processing activity of the native Apaf-1/caspase-9 apoptosome complex. J Biol Chem 279:19665–19682
Hill MM, Adrain C, Duriez PJ, Creagh EM, Martin SJ (2004) Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes. EMBO J 23:2134–2145
Yuan S, Topf M, Reubold TF, Eschenburg S, Akey CW (2013) Changes in apaf-1 conformation that drive apoptosome assembly. Biochemistry 52:2319–2327
Malladi S, Challa-Malladi M, Fearnhead HO, Bratton SB (2009) The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J 28:1916–1925
Li P, Nijhawan D, Budihardjo I et al (1997) Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91:479–489
Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y (1999) Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature 399:549–557
Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW (2010) Structure of an apoptosome-procaspase-9 CARD complex. Structure 18:571–583
Würstle ML, Laussmann MA, Rehm M (2012) The central role of initiator caspase-9 in apoptosis signal transduction and the regulation of its activation and activity on the apoptosome. Exp Cell Res 318:1213–1220
Yuan S, Yu X, Asara JM, Heuser JE, Ludtke SJ, Akey CW (2011) The holo-apoptosome: activation of procaspase-9 and interactions with caspase-3. Structure 19:1084–1096
Reubold TF, Eschenburg S (2012) A molecular view on signal transduction by the apoptosome. Cell Signal 24:1420–1425
Hu Q, Wu D, Chen W, Yan Z, Shi Y (2013) Proteolytic processing of the caspase-9 zymogen is required for apoptosome-mediated activation of caspase-9. J Biol Chem 288:15142–15147
Tinel A, Tschopp J (2004) The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304:843–846
Bouchier-Hayes L, Oberst A, McStay GP et al (2009) Characterization of cytoplasmic caspase-2 activation by induced proximity. Mol Cell 35:830–840
Jang TH, Zheng C, Wu H, Jeon JH, Park HH (2010) In vitro reconstitution of the interactions in the PIDDosome. Apoptosis 15:1444–1452
Janssens S, Tinel A (2012) The PIDDosome, DNA-damage-induced apoptosis and beyond. Cell Death Differ 19:13–20
Park HH, Logette E, Raunser S et al (2007) Death domain assembly mechanism revealed by crystal structure of the? Oligomeric PIDDosome core complex. Cell 128:533–546
Janssens S, Tinel A, Lippens S, Tschopp J (2005) PIDD mediates NF-kappaB activation in response to DNA damage. Cell 123:1079–1092
Tinel A, Janssens S, Lippens S et al (2007) Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. EMBO J 26:197–208
Ando K, Kernan JL, Liu PH et al (2012) PIDD death-domain phosphorylation by ATM controls Prodeath versus prosurvival PIDDosome signaling. Mol Cell 47:681–693
Manzl C, Krumschnabel G, Bock F et al (2009) Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol 185:291–303
Kim IR, Murakami K, Chen NJ et al (2009) DNA damage- and stress-induced apoptosis occurs independently of PIDD. Apoptosis 14:1039–1049
Vakifahmetoglu H, Olsson M, Orrenius S, Zhivotovsky B (2006) Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene 25:5683–5692
Imre G, Heering J, Takeda A-N et al (2012) Caspase-2 is an initiator caspase responsible for pore-forming toxin-mediated apoptosis. EMBO J 31:2615–2628
Jin Z, El-Deiry WS (2005) Overview of cell death signaling pathways. Cancer Biol Ther 4:139–163
Siegel RM, Frederiksen JK, Zacharias DA et al (2000) Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science 288:2354–2357
Siegel RM, Chan FK, Chun HJ, Lenardo MJ (2000) The multifaceted role of Fas signaling in immune cell homeostasis and autoimmunity. Nat Immunol 1:469–474
O’Reilly LA, Tai L, Lee L et al (2009) Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature 461:659–663
Lavrik IN (2014) Systems biology of death receptor networks: live and let die. Cell Death Dis 5:e1259
Scaffidi C, Fulda S, Srinivasan A et al (1998) Two CD95 (APO-1/Fas) signaling pathways. EMBO J 17:1675–1687
Yin XM, Wang K, Gross A et al (1999) Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature 400:886–891
Vercammen D, Beyaert R, Denecker G et al (1998) Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med 187:1477–1485
Holler N, Zaru R, Micheau O et al (2000) Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol 1:489–495
Chan FK-M, Shisler J, Bixby JG et al (2003) A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J Biol Chem 278:51613–51621
Adams JM, Cory S (1998) The Bcl-2 protein family: arbiters of cell survival. Science 281:1322–1326
Czabotar PE, Lessene G, Strasser A, Adams JM (2014) Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 15:49–63
Gupta S, Kass GEN, Szegezdi E, Joseph B (2009) The mitochondrial death pathway: a promising therapeutic target in diseases. J Cell Mol Med 13:1004–1033
Kulikov AV, Shilov ES, Mufazalov IA, Gogvadze V, Nedospasov SA, Zhivotovsky B (2012) Cytochrome c: the Achilles’ heel in apoptosis. Cell Mol Life Sci 69:1787–1797
Zamzami N, Kroemer G (2001) The mitochondrion in apoptosis: how Pandora’s box opens. Nat Rev Mol Cell Biol 2:67–71
Kerr JFR, Wyllie AH (1972) CAR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26:239–257
Akbari-Birgani S, Hosseinkhani S, Mollamohamadi S, Baharvand H (2014) Delay in apoptosome formation attenuates apoptosis in mouse. Embryonic stem cell differentiation. J Biol Chem 289:16905–16913
D’Brot A, Chen P, Vaishnav M, Yuan S, Akey CW, Abrams JM (2013) Tango7 directs cellular remodeling by the Drosophila apoptosome. Genes Dev 27:1650–1655
Favaloro B, Allocati N, Graziano V, Di Ilio C, De Laurenzi V (2012) Role of apoptosis in disease. Aging (Albany NY) 4:330–349
Yu X, Deng Q, Bode AM, Dong Z, Cao Y (2013) The role of necroptosis, an alternative form of cell death, in cancer therapy. Expert Rev Anticancer Ther 13:883–893
Bouralexis S, Findlay DM, Evdokiou A (2005) Death to the bad guys: targeting cancer via Apo2L/TRAIL. Apoptosis 10:35–51
Humphreys RC, Halpern W (2008) Trail receptors: targets for cancer therapy. Adv Exp Med Biol 615:127–158
Stegehuis JH, de Wilt LH, de Vries EGE, Groen HJ, de Jong S, Kruyt FE (2010) TRAIL receptor targeting therapies for non-small cell lung cancer: current status and perspectives. Drug Resist Update 13:2–15
Samali A, Szegezdi E, Mahalingham D et al (2009) Trail variants for treating cancer. WO2009077857-A2
Rosen GD (2000) Combined therapy of diterpenoid triepoxides and trail for synergistic killing of tumor cells. WO2000048619-A1
Zhao Y, Butler EB, Tan M (2013) Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis 4:e532
Kurokawa M, Kim J, Geradts J et al (2013) A network of substrates of the E3 ubiquitin ligases MDM2 and HUWE1 control apoptosis independently of p53. Sci Signal 6:ra32
Queudeville M, Seyfried F, Eckhoff SM et al (2012) Rapid engraftment of human ALL in NOD/SCID mice involves deficient apoptosis signaling. Cell Death Dis 3:e364
Hector S, Conlon S, Schmid J et al (2012) Apoptosome-dependent caspase activation proteins as prognostic markers in Stage II and III colorectal cancer. Br J Cancer 106:1499–1505
Chen W, Wang Q, Bai L et al (2014) RIP1 maintains DNA integrity and cell proliferation by regulating PGC-1α-mediated mitochondrial oxidative phosphorylation and glycolysis. Cell Death Differ 21:1061–1070
Johansson B, Mertens F, Mitelman F (1993) Cytogenetic deletion maps of hematologic neoplasms: circumstantial evidence for tumor suppressor loci. Genes Chromosom Cancer 8:205–218
Nam JY, Jong WL, Yun JK et al (2004) Loss of caspase-2, -6 and -7 expression in gastric cancers. APMIS 112:330–335
Holleman A, Den Boer ML, Kazemier KM et al (2005) Decreased PARP and procaspase-2 protein levels are associated with cellular drug resistance in childhood acute lymphoblastic leukemia. Blood 106:1817–1823
Acknowledgments
The work in the authors group was supported by grants from the Russian Science Foundation (14-25-00056), Russian Foundation for Basic Research, Russian President Fund, Dynasty Foundation, as well as the Stockholm Cancer Societies, the Swedish Childhood Cancer Foundation, the Swedish Research Council.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zamaraev, A.V., Kopeina, G.S., Zhivotovsky, B. et al. Cell death controlling complexes and their potential therapeutic role. Cell. Mol. Life Sci. 72, 505–517 (2015). https://doi.org/10.1007/s00018-014-1757-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-014-1757-2