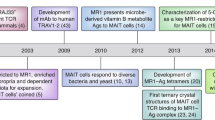
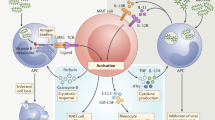
Abstract
Mucosa-associated invariant T (MAIT) cells are a unique population of innate T cells that are abundant in humans. These cells possess an evolutionarily conserved invariant T cell receptor α chain restricted by the nonpolymorphic class Ib major histocompatibility (MHC) molecule, MHC class I-related protein (MR1). The recent discovery that MAIT cells are activated by MR1-bound riboflavin metabolite derivatives distinguishes MAIT cells from all other αβ T cells in the immune system. Since mammals lack the capacity to synthesize riboflavin, intermediates from the riboflavin biosynthetic pathway are distinct microbial molecular patterns that provide a unique signal to the immune system. Multiple lines of evidence suggest that MAIT cells, which produce important cytokines such as IFN-γ, TNF, and IL-17A, have the potential to influence immune responses to a broad range of pathogens. Here we will discuss our current understanding of MAIT cell biology and their role in pathogen defense.
Similar content being viewed by others
References
Medzhitov R, Janeway CA Jr (2002) Decoding the patterns of self and nonself by the innate immune system. Science 296(5566):298–300
Litman GW, Rast JP, Fugmann SD (2010) The origins of vertebrate adaptive immunity. Nat Rev Immunol 10(8):543–553
Hirano M, Das S, Guo P, Cooper MD (2011) The evolution of adaptive immunity in vertebrates. Adv Immunol 109:125–157
Von Boehmer H (1991) Positive and negative selection of the alpha beta T cell repertoire in vivo. Curr Opin Immunol 3(2):210–215
Lawson V (2012) Turned on by danger: activation of CD1d-restricted invariant natural killer T cells. Immunology 137(1):20–27
Porcelli S, Yockey CE, Brenner MB, Balk SP (1993) Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4− CD8− α/β T cells demonstrates preferential use of several Vβ genes and an invariant TCR α chain. J Exp Med 178(1):1–16
Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB (1004) Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature 372(6507):691–694
Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, Bendelac A, Bonneville M, Lantz O (1999) An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med 189(12):1907–1921
Gold MC, McLaren JE, Reistetter JA, Smyk-Pearson S, Ladell K, Swarbick GM, Yu YYL, Hansen TH, Lund O, Nielsen M, Gerritsen B, Kesmir C, Miles JJ, Lewinsohn DA, Price DA, Lewinsohn DM (2014) MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J Exp Med 211(8):1601–1610
Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, Eckle SBG, Uldrich AP, Birkinshaw RW, Patel O, Kostenko L, Meehan B, Kedziereska K, Liu L, Fairlie DP, Hansen TH, Godfrey DI, Rossjohn J, McCluskey J, Kjer-Nielsen L (2013) Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med 210(11):2305–2320
Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, Williamson NA, Strugnell RA, Van Sinderen D, Mak JY, Fairlie DP, Kjer-Nielsen L, Rossjohn J, McCluskey J (2014) T cell activation by transitory neo-antigens from distinct microbial pathways. Nature 509(7500):361–365
Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O’Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J (2012) MR1 presents vitamin B metabolites to MAIT cells. Nature 491(7426):717–723
Riegert P, Wanner V, Bahram S (1999) Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J Immunol 161980:4066–4077
Tsukamoto K, Deakin JE, Graves JA, Hashimoto K (2013) Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics 65(2):115–124
Yamaguchi H, Kurosawa Y, Hashimoto K (1998) Expanded genomic organization of conserved mammalian MHC class I-related genes, human MR1 and its murine ortholog. Biochem Biophys Res Commun 250(3):558–564
Hansen TH, Huang S, Arnold PL, Fremont DH (2007) Patterns of nonclassical MHC antigen presentation. Nat Immunol 8(6):563–568
Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, Winata E, Swarbrick GM, Chua WJ, Yu YY, Lantz O, Cook MS, Null MD, Jacoby DB, Harriff MJ, Lewinsohn DA, Hansen TH, Lewinsohn DM (2010) Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol 8(6):e1000407
Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, Levy E, Dusseaux M, Meyssonnier V, Premel V, Ngo C, Riteau B, Duban L, Robert D, Huang S, Rottman M, Soudais C, Lantz O (2010) Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol 11(8):701–708
Chua WJ, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, Hansen TH (2012) Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun 80(9):3256–3267
Meierovics A, Yankelevich WJ, Cowley SC (2013) MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci USA 110(33):E119–E128
Brigl M, Brenner MB (2010) How invariant natural killer T cells respond to infection by recognizing microbial of endogenous antigens. Semin Immunol 22(2):79–86
Brigl M, Tatituri RV, Watts GF, Bhowruth V, Leadbetter EA, Barton N, Cohen NR, Hsu FF, Besra GS, Brenner MB (2011) Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med 208(6):1163–1177
Billerbeck E, Kanh YH, Walker L, Lockstone H, Grafmueller S, Fleming V, Flint J, Willberg CB, Bengsch B, Seigel B, Ramamurthy N, Zitzmann N, Barnes EJ, Thevanayagam J, Bhagwanani A, Leslie A, Oo YH, Kollnberger S, Bowness P, Drognitz O, Adams DH, Blum HE, Thimme R, Klenerman P (2010) Analysis of CD161 expression of human CD8+ T cells defines a distinct functional subset with tissue-homing properties. Proc Natl Acad Sci USA 107(7):3006–3011
Ussher JE, Bilton M, Attwod E, Shadwell J, Richardson R, de Lara C, Mettke E, Kurioka A, Hansen TH, Klenerman P, Willberg CB (2014) CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12 + IL-18 in a TCR-independent manner. Eur J Immunol 44(1):195–203
Le Bourhis L, Dusseaux M, Bohineust A, Bessoles S, Martin E, Premel V, Core M, Sleurs D, Serriari NE, Treiner E, Hovroz C, Sansonetti P, Gougeon ML, Soudais C, Lantz O (2013) MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog 9(10):e1003681
Salerno-Goncalves R, Rezwan T, Sztein MB (2014) B cells modulate mucosal associated invariant T cell immune responses. Front Immunol 4(511):1–15
Huang S, Gilfillan S, Kim S, Thompson B, Wang X, Sant AJ, Fremont DH, Lantz O, Hansen TH (2008) MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J Exp Med 205(5):1201–1211
Annibali V, Ristori G, Angelini DF, Serafini B, Mechelli R, Cannoni S, Romano S, Paolillo A, Abderrahim H, Diamantini A, Borsellino G, Aloisi F, Battistini L, Salvetti M (2011) CD161high CD8+ T cells bear pathogenic potential in multiple sclerosis. Brain 134(pt 2):542–554
Croxford JL, Miyake S, Huang Y, Shimamura M, Yamamura T (2006) Invariant Vα19i T cells regulate autoimmune inflammation. Nat Immunol 7(9):987–994
Chiba A, Tajima R, Tomi C, Miyazaki Y, Yamamura T, Miyake S (2012) Mucosal-associated invariant T cells promote inflammation and exacerbate disease in murine models of arthritis. Arthritis Rheum 64(1):153–161
Treiner E, Durban L, Bahram S, Radosacljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O (2003) Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422(6928):160–164
Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, Louis D, Milder M, Le Bourhis L, Soudais C, Treiner E, Lantz O (2011) Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting cells. Blood 117(4):1250–1259
Lepore M, Kalinichenko A, Colone A, Paleja B, Singhal A, Tschumi A, Lee B, Poidinger M, Zolezzi F, Quagliata L, Sander P, Newell E, Bertoletti A, Terracciano L, De Libero G, Mori L (2014) Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. Nat Commun 5:3866. doi:10.1038/ncomms4866
Grimaldi D, Le Bourhis L, Sauneuf B, Dechartes A, Rousseau C, Ouaaz F, Milder M, Louis D, Chiche J, Mira J, Lantz O, Pene F (2014) Specific MAIT behaviour among innate-like lymphocytes in critically ill patients with severe infections. Intensive Care Med 40(2):192–201
Georgel P, Radosalvljevic M, Macquin C, Bahram S (2011) The non-conventional MHC class I MR1 molecule controls infection by Klebsiella pneumoniae in mice. Mol Immunol 48(5):769–775
Schneeman M, Schoeden G (2007) Macrophage biology and immunology: man is not a mouse. J Infect Dis 81(3):579
Walker LJ, Kang YH, Smith MO, Tharmalingham H, Ramamurthy N, Fleming VM, Sahgal N, Leslie A, Oo Y, Geremia A, Scriba TJ, Hanekom WA, Lauer GM, Lantz O, Adams DH, Powrie F, Barnes E, Klenerman P (2012) Human MAIT and CD8αα cells develop from a pool of type-17 precommitted CD8+ T cells. Blood 119(2):422–433
Gold MC, Eid T, Smyk-Pearson S, Eberling Y, Swarbrick GM, Langley SM, Streeter PR, Lewinsohn DA, Lewinsohn DM (2013) Human thymic MR1-restricted MAIT cells are innate pathogen-reactive effectors that adapt following thymic egress. Mucosal Immunol 6(1):35–44
Colmone A, Wang C (2006) H2-M3-restricted T cell response to infection. Microbes Infect 8(8):2277–2283
Sandberg JK, Dias J, Shacklett BJ, Leeansyah E (2013) Will loss of your mucosa-associated invariant T cells weaken your HAART? AIDS 27(16):2501–2504
Leeansyah E, Ganesh A, Quigley MF, Sonnerborg A, Andersson J, Hunt PW, Somsouk M, Deeks SG, Martin JN, Moll M, Shacklett BL, Sandberg JK (2013) Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood 121(7):1124–1135
Cosgrove C, Ussher JE, Rauch A, Gartner K, Kurioka A, Huhn MH, Adelman K, Kang YH, Fergusson JR, Simmonds P, Goulder P, Hansen TH, Fox J, Gunthard HF, Khanna N, Powrie F, Steel A, Gazzard B, Phillips RE, Frater J, Uhlig H, Klenerman P (2013) Early and nonreversible decrease of CD161++/MAIT cells in HIV infection. Blood 121(6):951–961
Greathead L, Metcalf R, Gazzard B, Gotch F, Steel A, Kelleher P (2014) CD8+/CD161++ mucosal-associated invariant T-cell levels in the colon are restored on long-term antiretroviral therapy and correlate with CD8+ T-cell immune activation. AIDS 28(11):1690–1692
Wong EB, Akilimali NA, Govender P, Sullivan ZA, Cosgrove C, Pillay M, Lewinsohn DM, Bishai WR, Walker BD, Ndung’u T, Klenerman P, Kasprowicz VO (2013) Low levels of peripheral CD161++ CD8+ mucosal associated invariant T (MAIT) cells are found in HIV and HIV/TB co-infection. PLoS ONE 8(12):e83474
Sonnenberg P, Glynn JR, Fielding K, Murray J, Godfrey-Faussett P, Shearer S (2005) How soon after infection with HIV does the risk of tuberculosis start to increase? A retrospective cohort study in South African gold miners. J Infect Dis 191(2):150–158
Le Bourhis L, Mburu YK, Lantz O (2013) MAIT cells, surveyors of a new class of antigen: development and functions. Curr Opin Immunol 25(2):174–180
Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A (2008) The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29(3):391–403
Leeansyah E, Loh L, Nixon DF, Sandberg JK (2014) Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat Commun 5(3143):1–10
Bienemann K, Iouannidou K, Schoenberg K, Krux F, Reuther S, Feyen O, Bienneman K, Schuster F, Uhrburg M, Laws HJ, Borkhardt A (2011) iNKT cell frequency in peripheral blood of Caucasian children and adolescent: the absolute iNKT cell count is stable from birth to adulthood. Scand J Immunol 74(4):406–411
Ostantin DV, Brown CM, Gray L, Bharwani S, Grisham MB (2010) Evaluation of the immunoregulatory activity of intraepithelial lymphocytes in a mouse model of chronic intestinal inflammation. Int Immunol 22(12):927–939
Kawachi I, Maldonado J, Strader C, Gilfillan S (2006) MR1-restricted Vα19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J Immunol 176(3):1618–1627
Zeissig S, Blumberg RS (2013) Commensal microbiota and NKT cells in the control of inflammatory diseases at mucosal surfaces. Curr Opin Immunol 25(6):690–696
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cowley, S.C. MAIT cells and pathogen defense. Cell. Mol. Life Sci. 71, 4831–4840 (2014). https://doi.org/10.1007/s00018-014-1708-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-014-1708-y