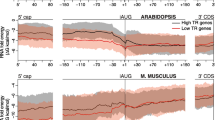
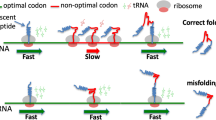
Abstract
Quantitative control of gene expression occurs at multiple levels, including the level of translation. Within the overall process of translation, most identified regulatory processes impinge on the initiation phase. However, recent studies have revealed that the elongation phase can also regulate translation if elongation and initiation occur with specific, not mutually compatible rate parameters. Translation elongation then limits the overall amount of protein that can be made from an mRNA. Several recently discovered control mechanisms of biological pathways are based on such elongation control. Here, we review the molecular mechanisms that determine ribosome speed in eukaryotic organisms, and discuss under which conditions ribosome speed can become the controlling parameter of gene expression levels.





Similar content being viewed by others
References
Hershey JWB, Sonenberg N, Mathews M (2007) Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press, Woodbury NY
Aitken CE, Lorsch JR (2012) A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 19:568–576. doi:10.1038/nsmb.2303
von der Haar T, Valášek LS (2014) mRNA Translation: Fungal Variations on a Eukaryotic Theme Tobias. In: Sesma A, von der Haar T (eds) Fungal RNA Biol. Springer International Publishing, Heidelberg, pp 113–134
Richter JD, Sonenberg N (2005) Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433:477–480. doi:10.1038/nature03205
MacDonald CT, Gibbs JH, Pipkin AC (1968) Kinetics of biopolymerization on nucleic acid templates. Biopolymers 6:1–25. doi:10.1002/bip.1968.360060102
Gordon R (1969) Polyribosome dynamics at steady state. J Theor Biol 22:515–532. doi:10.1016/0022-5193(69)90018-6
Heinrich R, Rapoport TA (1980) Mathematical modelling of translation of mRNA in eucaryotes; steady state, time-dependent processes and application to reticulocytes. J Theor Biol 86:279–313. doi:10.1016/0022-5193(80)90008-9
Zhou M, Guo J, Cha J et al (2013) Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495:111–115. doi:10.1038/nature11833
Chan CTY, Pang YLJ, Deng W et al (2012) Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 3:937. doi:10.1038/ncomms1938
Kemp AJ, Betney R, Ciandrini L et al (2013) A yeast tRNA mutant that causes pseudohyphal growth exhibits reduced rates of CAG codon translation. Mol Microbiol 87:284–300. doi:10.1111/mmi.12096
Chu D, Kazana E, Bellanger N et al (2014) Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J 33:21–34. doi:10.1002/embj.201385651
Phizicky EM, Hopper AK (2010) tRNA biology charges to the front. Genes Dev 24:1832–1860. doi:10.1101/gad.1956510
Boguta M (2013) Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta 1829:376–384. doi:10.1016/j.bbagrm.2012.11.004
Tavenet A, Suleau A, Dubreuil G et al (2009) Genome-wide location analysis reveals a role for Sub1 in RNA polymerase III transcription. Proc Natl Acad Sci USA 106:14265–14270. doi:10.1073/pnas.0900162106
Candelas GC, Arroyo G, Carrasco C, Dompenciel R (1990) Spider silkglands contain a tissue-specific alanine tRNA that accumulates in vitro in response to the stimulus for silk protein synthesis. Dev Biol 140:215–220. doi:10.1016/0012-1606(90)90069-U
Cintron I, Capo L, Plazaola A et al (1999) A spider tRNA(Ala) requires a far upstream sequence element for expression. Gene 132:195–201
Ikemura T (1982) Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer R. J Mol Biol 158:573–597. doi:10.1016/0022-2836(82)90250-9
Hani J, Feldmann H (1998) tRNA genes and retroelements in the yeast genome. Nucleic Acids Res 26:689–696. doi:10.1093/nar/26.3.689
Bloom-Ackermann Z, Navon S, Gingold H et al (2014) A comprehensive tRNA deletion library unravels the genetic architecture of the tRNA pool. PLoS Genet 10:e1004084. doi:10.1371/journal.pgen.1004084
Dittmar KA, Goodenbour JM, Pan T (2006) Tissue-specific differences in human transfer RNA expression. PLoS Genet 2:e221. doi:10.1371/journal.pgen.0020221
Schlegel RA, Iversen P, Rechsteiner M (1978) The turnover of tRNAs microinjected into animal cells. Nucleic Acids Res 5:3715–3729. doi:10.1093/nar/5.10.3715
Hopper AK (2013) Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194:43–67. doi:10.1534/genetics.112.147470
Alexandrov A, Chernyakov I, Gu W et al (2006) Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21:87–96. doi:10.1016/j.molcel.2005.10.036
Thompson DM, Lu C, Green PJ, Parker R (2008) tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14:2095–2103. doi:10.1261/rna.1232808
Saxena SK, Sirdeshmukh R, Ardelt W et al (2002) Entry into cells and selective degradation of tRNAs by a cytotoxic member of the RNase A family. J Biol Chem 277:15142–15146. doi:10.1074/jbc.M108115200
Jablonowski D, Schaffrath R (2007) Zymocin, a composite chitinase and tRNase killer toxin from yeast. Biochem Soc Trans 35:1533–1537. doi:10.1042/BST0351533
Kirsebom LA (2007) RNase P RNA mediated cleavage: substrate recognition and catalysis. Biochimie 89:1183–9114. doi:10.1016/j.biochi.2007.05.009
Vogel A, Schilling O, Späth B, Marchfelder A (2005) The tRNAse Z family of proteins: physiological functions, substrate specificity and structural properties. Biol Chem 386:1253–1264
Betat H, Rammelt C, Mörl M (2010) tRNA nucleotidyltransferases: ancient catalysts with an unusual mechanism of polymerization. Cell Mol Life Sci 67:1447–1463. doi:10.1007/s00018-010-0271-4
Popow J, Schleiffer A, Martinez J (2012) Diversity and roles of (t)RNA ligases. Cell Mol Life Sci 69:2657–2670. doi:10.1007/s00018-012-0944-2
El Yacoubi B, Lyons B, Cruz Y et al (2009) The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res 37:2894–2909. doi:10.1093/nar/gkp152
Dreher TW, Uhlenbeck OC, Browning KS (1999) Quantitative Assessment of EF-1α GTP Binding to Aminoacyl-tRNAs, Aminoacyl-viral RNA, and tRNA Shows Close Correspondence to the RNA Binding Properties of EF-Tu. J Biol Chem 274:666–672. doi:10.1074/jbc.274.2.666
Ling J, Reynolds NM, Ibba M (2009) Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol 63:61–78
Qian W, Yang J-R, Pearson NM et al (2012) Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet 8:e1002603. doi:10.1371/journal.pgen.1002603
McLaughlin CS, Magee PT, Hartwell LH (1969) Role of isoleucyl-transfer ribonucleic acid synthetase in ribonucleic acid synthesis and enzyme repression in yeast. J Bacteriol 100:579–584
Messenguy F, Delforge J (1976) Role of transfer ribonucleic acids in the regulation of several biosyntheses in Saccharomyces cerevisiae. Eur J Biochem 67:335–339. doi:10.1111/j.1432-1033.1976.tb10696.x
Johansson MJO, Esberg A, Huang B et al (2008) Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol 28:3301–3312. doi:10.1128/MCB.01542-07
Chu D, Barnes DJ, von der Haar T (2011) The role of tRNA and ribosome competition in coupling the expression of different mRNAs in Saccharomyces cerevisiae. Nucleic Acids Res 39:6705–6714. doi:10.1093/nar/gkr300
Rodnina MV, Wintermeyer W (2009) Recent mechanistic insights into eukaryotic ribosomes. Curr Opin Cell Biol 21:435–443. doi:10.1016/j.ceb.2009.01.023
Pape T, Wintermeyer W, Rodnina M (1999) Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J 18:3800–3807. doi:10.1093/emboj/18.13.3800
Gromadski KB, Rodnina MV (2004) Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell 13:191–200
Fluitt A, Pienaar E, Viljoen H (2007) Ribosome kinetics and aa-tRNA competition determine rate and fidelity of peptide synthesis. Comput Biol Chem 31:335–346. doi:10.1016/j.compbiolchem.2007.07.003
Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2:13–34
Bulmer M (1987) Coevolution of codon usage and transfer RNA abundance. Nature 325:728–730. doi:10.1038/325728a0
Kramer EB, Vallabhaneni H, Mayer LM, Farabaugh PJ (2010) A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA 16:1797–1808. doi:10.1261/rna.2201210
Plant EP, Nguyen P, Russ JR et al (2007) Differentiating between near- and non-cognate codons in Saccharomyces cerevisiae. PLoS ONE 2:e517. doi:10.1371/journal.pone.0000517
Curran JF (1995) Decoding with the A: I wobble pair is inefficient. Nucleic Acids Res 23:683–688. doi:10.1093/nar/23.4.683
Björk GR, Huang B, Persson OP, Byström AS (2007) A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13:1245–1255. doi:10.1261/rna.558707
Kothe U, Rodnina MV (2007) Codon reading by tRNAAla with modified uridine in the wobble position. Mol Cell 25:167–174. doi:10.1016/j.molcel.2006.11.014
Stadler M, Fire A (2011) Wobble base-pairing slows in vivo translation elongation in metazoans. RNA 17:2063–2073. doi:10.1261/rna.02890211
Cannarrozzi G, Schraudolph NN, Faty M et al (2010) A role for codon order in translation dynamics. Cell 141:355–367. doi:10.1016/j.cell.2010.02.036
McGuffee SR, Elcock AH (2010) Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput Biol 6:e1000694. doi:10.1371/journal.pcbi.1000694
Mirande M (2010) Processivity of translation in the eukaryote cell: role of aminoacyl-tRNA synthetases. FEBS Lett 584:443–447. doi:10.1016/j.febslet.2009.11.027
Bulova SI, Burka ER (1970) Biosynthesis of nonglobin protein by membrane-bound ribosomes in reticulocytes. J Biol Chem 245:4907–4912
Vedeler A, Pryme IF, Hesketh JE (1991) The characterization of free, cytoskeletal and membrane-bound polysomes in Krebs II ascites and 3T3 cells. Mol Cell Biochem 100:183–193
Kopeina GS, Afonina ZA, Gromova KV et al (2008) Step-wise formation of eukaryotic double-row polyribosomes and circular translation of polysomal mRNA. Nucleic Acids Res 36:2476–2488. doi:10.1093/nar/gkm1177
Brandt F, Carlson L-A, Hartl FU et al (2010) The three-dimensional organization of polyribosomes in intact human cells. Mol Cell 39:560–569. doi:10.1016/j.molcel.2010.08.003
Pfeffer S, Brandt F, Hrabe T et al (2012) Structure and 3D arrangement of endoplasmic reticulum membrane-associated ribosomes. Structure 20:1508–1518. doi:10.1016/j.str.2012.06.010
Pavlov MY, Watts RE, Tan Z et al (2009) Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci USA 106:50–54. doi:10.1073/pnas.0809211106
Gutierrez E, Shin B-S, Woolstenhulme CJ et al (2013) eIF5A promotes translation of polyproline motifs. Mol Cell 51:35–45. doi:10.1016/j.molcel.2013.04.021
Johansson M, Ieong K-W, Trobro S et al (2011) pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc Natl Acad Sci USA 108:79–84. doi:10.1073/pnas.1012612107
Zinshteyn B, Gilbert WV (2013) Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling. PLoS Genet 9:e1003675. doi:10.1371/journal.pgen.1003675
Lareau LF, Hite DH, Hogan GJ, Brown PO (2014) Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife 3:e01257. doi:10.7554/eLife01257
Charneski CA, Hurst LD (2013) Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol 11:e1001508. doi:10.1371/journal.pbio.1001508
Lu J, Deutsch C (2008) Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol 384:73–86. doi:10.1016/j.jmb.2008.08.089
Tuller T, Veksler-Lublinsky I, Gazit N et al (2011) Composite effects of gene determinants on the translation speed and density of ribosomes. Genome Biol 12:R110. doi:10.1186/gb-2011-12-11-r110
Tenson T, Ehrenberg M (2002) Regulatory nascent peptides in the ribosomal tunnel. Cell 108:591–594
Horton LE, James P, Craig EA, Hensold JO (2001) The yeast hsp70 homologue Ssa is required for translation and interacts with Sis1 and Pab1 on translating ribosomes. J Biol Chem 276:14426–14433. doi:10.1074/jbc.M100266200
Gautschi M, Lilie H, Fünfschilling U et al (2001) RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci USA 98:3762–3767. doi:10.1073/pnas.071057198
Rakwalska M, Rospert S (2004) The Ribosome-Bound Chaperones RAC and Ssb1/2p are required for accurate translation in saccharomyces cerevisiae. Mol Cell Biol 24:9186–9197. doi:10.1128/MCB.24.20.9186
Willmund F, del Alamo M, Pechmann S et al (2013) The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152:196–209. doi:10.1016/j.cell.2012.12.001
Shalgi R, Hurt JA, Krykbaeva I et al (2013) Widespread regulation of translation by elongation pausing in heat shock. Mol Cell 49:439–452. doi:10.1016/j.molcel.2012.11.028
Chiabudini M, Conz C, Reckmann F, Rospert S (2012) Ribosome-associated complex and Ssb are required for translational repression induced by polylysine segments within nascent chains. Mol Cell Biol 32:4769–4779. doi:10.1128/MCB.00809-12
Hirschmann WD, Westendorf H, Mayer A, et al. (2014) Scp160p is required for translational efficiency of codon-optimized mRNAs in yeast. Nucleic Acids Res (in press). doi: 10.1093/nar/gkt1392
von Heijne G, Nilsson L, Blomberg C (1977) Translation and messenger RNA secondary structure. J Theor Biol 68:321–329. doi:10.1016/0022-5193(77)90063-7
Tuller T, Waldman YY, Kupiec M, Ruppin E (2010) Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA 107:3645–3650. doi:10.1073/pnas.0909910107
Zur H, Tuller T (2012) Strong association between mRNA folding strength and protein abundance in S. cerevisiae. EMBO Rep 13:272–277. doi:10.1038/embor.2011.262
Shaw LB, Zia RKP, Lee KH (2003) Totally asymmetric exclusion process with extended objects: a model for protein synthesis. Phys Rev E 68:1–17. doi:10.1103/PhysRevE.68.021910
Chou T, Lakatos G (2004) Clustered bottlenecks in mrna translation and protein synthesis. Phys Rev Lett 93:1–4. doi:10.1103/PhysRevLett93198101
Dong JJ, Schmittmann B, Zia RKP (2006) Towards a model for protein production rates. J Stat Phys 128:21–34. doi:10.1007/s10955-006-9134-7
Ciandrini L, Stansfield I, Romano MC (2013) Ribosome traffic on mRNAs maps to gene ontology: genome-wide quantification of translation initiation rates and polysome size regulation. PLoS Comput Biol 9:e1002866. doi:10.1371/journal.pcbi.1002866
Mao Y, Liu H, Liu Y, Tao S (2014) Deciphering the rules by which dynamics of mRNA secondary structure affect translation efficiency in Saccharomyces cerevisiae. Nucleic Acids Res 42:4813–4822. doi:10.1093/nar/gku159
Tuller T, Carmi A, Vestsigian K et al (2010) An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141:344–354. doi:10.1016/j.cell.2010.03.031
Pechmann S, Frydman J (2013) Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol 20:237–243. doi:10.1038/nsmb.2466
Bentele K, Saffert P, Rauscher R et al (2013) Efficient translation initiation dictates codon usage at gene start. Mol Syst Biol 9:675. doi:10.1038/msb.2013.32
Ingolia NT (2014) Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet 1–9
Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147:789–802. doi:10.1016/j.cell.2011.10.002
Li G-W, Oh E, Weissman JS (2012) The anti-Shine–Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484:538–541. doi:10.1038/nature10965
Dana A, Tuller T (2012) Determinants of translation elongation speed and ribosomal profiling biases in mouse embryonic stem cells. PLoS Comput Biol 8:e1002755. doi:10.1371/journal.pcbi.1002755
Letzring DP, Dean KM, Grayhack EJ (2010) Control of translation efficiency in yeast by codon-anticodon interactions. RNA 16:2516–2528. doi:10.1261/rna.2411710
von der Haar T (2008) A quantitative estimation of the global translational activity in logarithmically growing yeast cells. BMC Syst Biol 2:87. doi:10.1186/1752-0509-2-87
Chu D, von der Haar T (2012) The architecture of eukaryotic translation. Nucleic Acids Res 40:10098–10106. doi:10.1093/nar/gks825
Shah P, Ding Y, Niemczyk M et al (2013) Rate-limiting steps in yeast protein translation. Cell 153:1589–1601. doi:10.1016/j.cell.2013.05.049
Shah P, Gilchrist MA (2011) Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift. Proc Natl Acad Sci USA 108:10231–10236. doi:10.1073/pnas.1016719108
Firczuk H, Kannambath S, Pahle J et al (2013) An in vivo control map for the eukaryotic mRNA translation machinery. Mol Syst Biol 9:1–13. doi:10.1038/msb.2012.73
Warner JR, Knopf PM, Rich A (1963) A multiple ribosomal structure in protein synthesis. Proc Natl Acad Sci USA 49:122–129
MacDonald CT, Gibbs JH (1969) Concerning the kinetics of polypeptide synthesis on polyribosomes. Biopolymers 7:707–725. doi:10.1002/bip.1969.360070508
Mahlab S, Linial M (2014) Speed controls in translating secretory proteins in eukaryotes - an evolutionary perspective. PLoS Comput Biol 10:e1003294. doi:10.1371/journal.pcbi.1003294
Rapoport TA, Heinrich R, Walter P, Schulmeister T (1987) Mathematical modeling of the effects of the signal recognition particle on translation and translocation of proteins across the endoplasmic reticulum membrane. J Mol Biol 195:621–636. doi:10.1016/0022-2836(87)90186-0
Neafsey DE, Galagan JE (2007) Positive selection for unpreferred codon usage in eukaryotic genomes. BMC Evol Biol 7:119
Drummond DA, Wilke CO (2008) Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134:341–352. doi:10.1016/j.cell.2008.05.042
Thanaraj TA, Argos P (1996) Protein secondary structural types are differentially coded on messenger RNA. Protein Sci 5:1973–1983. doi:10.1002/pro.5560051003
Saunders R, Deane CM (2010) Synonymous codon usage influences the local protein structure observed. Nucleic Acids Res 38:6719–6728. doi:10.1093/nar/gkq495
Buchan JR, Aucott LS, Stansfield I (2006) tRNA properties help shape codon pair preferences in open reading frames. Nucleic Acids Res 34:1015–1027. doi:10.1093/nar/gkj488
Trotta E (2013) Selection on codon bias in yeast: a transcriptional hypothesis. Nucleic Acids Res 41:9382–9395. doi:10.1093/nar/gkt740
Comeron JM, Kreitman M, Aguade M (1999) Natural Selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics 151:239–249
Stergachis AB, Haugen E, Shafer A et al (2013) Exonic transcription factor binding directs codon choice and affects protein evolution. Science 342(80):1367–1372
Nakagawa S, Niimura Y, Gojobori T et al (2008) Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res 36:861–871. doi:10.1093/nar/gkm1102
Zur H, Tuller T (2013) New universal rules of eukaryotic translation initiation fidelity. PLoS Comput Biol 9:e1003136. doi:10.1371/journal.pcbi.1003136
Acknowledgments
TvdH acknowledges support for work relevant to the topic of this review from the Royal Society, UK (RG090785) and the Biotechnology and Biological Sciences Research Council, UK (I010351).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tarrant, D., von der Haar, T. Synonymous codons, ribosome speed, and eukaryotic gene expression regulation. Cell. Mol. Life Sci. 71, 4195–4206 (2014). https://doi.org/10.1007/s00018-014-1684-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-014-1684-2