Abstract
DNA methylation has been studied comprehensively and linked to both normal neurodevelopment and neurological diseases. The recent identification of several new DNA modifications, including 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine, has given us a new perspective on the previously observed plasticity in 5mC-dependent regulatory processes. Here, we review the latest research into these cytosine modifications, focusing mainly on their roles in neurodevelopment and diseases.

Similar content being viewed by others
References
Holliday R (2006) Epigenetics: a historical overview. Epigenetics 1(2):76–80
Waddington CH (1939) An introduction to modern genetics. G. Allen & Unwin, London
Schreiber SL, Bernstein BE (2002) Signaling network model of chromatin. Cell 111(6):771–778
Dupont C, Armant DR, Brenner CA (2009) Epigenetics: definition, mechanisms and clinical perspective. Semin Reprod Med 27(5):351–357. doi:10.1055/s-0029-1237423
Jaenisch R, Bird A (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33(Suppl):245–254. doi:10.1038/ng1089
Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3(6):415–428. doi:10.1038/nrg816
Chi P, Allis CD, Wang GG (2010) Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10(7):457–469. doi:10.1038/nrc2876
Reik W (2007) Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447(7143):425–432. doi:10.1038/nature05918
Griffith JS, Mahler HR (1969) DNA ticketing theory of memory. Nature 223(5206):580–582
Suzuki MM, Bird A (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9(6):465–476. doi:10.1038/nrg2341
Li E (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 3(9):662–673. doi:10.1038/nrg887
Gopalakrishnan S, Van Emburgh BO, Robertson KD (2008) DNA methylation in development and human disease. Mutat Res 647(1–2):30–38. doi:10.1016/j.mrfmmm.2008.08.006
Jakovcevski M, Akbarian S (2012) Epigenetic mechanisms in neurological disease. Nat Med 18(8):1194–1204. doi:10.1038/nm.2828
Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H (2010) Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci 13(11):1338–1344. doi:10.1038/nn.2672
Baylin SB, Jones PA (2011) A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 11(10):726–734. doi:10.1038/nrc3130
Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99(3):247–257
Bestor TH (2000) The DNA methyltransferases of mammals. Hum Mol Genet 9(16):2395–2402
Anderson RM, Bosch JA, Goll MG, Hesselson D, Dong PD, Shin D, Chi NC, Shin CH, Schlegel A, Halpern M, Stainier DY (2009) Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev Biol 334(1):213–223. doi:10.1016/j.ydbio.2009.07.017
Sun LQ, Lee DW, Zhang Q, Xiao W, Raabe EH, Meeker A, Miao D, Huso DL, Arceci RJ (2004) Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev 18(9):1035–1046. doi:10.1101/gad.1176104
Hendrich B, Bird A (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol 18(11):6538–6547
Bogdanovic O, Veenstra GJ (2009) DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma 118(5):549–565. doi:10.1007/s00412-009-0221-9
Lopez-Serra L, Esteller M (2008) Proteins that bind methylated DNA and human cancer: reading the wrong words. Br J Cancer 98(12):1881–1885. doi:10.1038/sj.bjc.6604374
Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY (2008) MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320(5880):1224–1229. doi:10.1126/science.1153252
Guy J, Cheval H, Selfridge J, Bird A (2011) The role of MeCP2 in the brain. Annu Rev Cell Dev Biol 27:631–652. doi:10.1146/annurev-cellbio-092910-154121
Wyatt GR, Cohen SS (1953) The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem J 55(5):774–782
Penn NW, Suwalski R, O’Riley C, Bojanowski K, Yura R (1972) The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem J 126(4):781–790
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324(5929):930–935. doi:10.1126/science.1170116
Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466(7310):1129–1133. doi:10.1038/nature09303
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333(6047):1303–1307. doi:10.1126/science.1210944
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333(6047):1300–1303. doi:10.1126/science.1210597
Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N (2012) MeCP2 Binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151(7):1417–1430. doi:10.1016/j.cell.2012.11.022
Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, Vasanthakumar A, Godley LA, Chang Q, Cheng X, He C, Jin P (2011) 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci 14(12):1607–1616. doi:10.1038/nn.2959
Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C (2011) Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol 29(1):68–72. doi:10.1038/nbt.1732
Kriaucionis S, Heintz N (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324(5929):929–930. doi:10.1126/science.1169786
Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T (2010) Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem 49(31):5375–5377. doi:10.1002/anie.201002033
Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A (2011) Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473(7347):394–397. doi:10.1038/nature10102
Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, Yoon YS, Ren B, He C, Jin P (2011) Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet 7(6):e1002154. doi:10.1371/journal.pgen.1002154
Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C (2012) Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149(6):1368–1380. doi:10.1016/j.cell.2012.04.027
Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G (2010) Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci 13(4):423–430. doi:10.1038/nn.2514
Miller CA, Sweatt JD (2007) Covalent modification of DNA regulates memory formation. Neuron 53(6):857–869. doi:10.1016/j.neuron.2007.02.022
Feng J, Chang H, Li E, Fan G (2005) Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res 79(6):734–746. doi:10.1002/jnr.20404
Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H (1994) Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation 56(1–2):39–44
Li E, Bestor TH, Jaenisch R (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69(6):915–926
Golshani P, Hutnick L, Schweizer F, Fan G (2005) Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Relat Syst 3(3):227–233. doi:10.1017/S1472928807000222
Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R (2007) Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn 236(6):1663–1676. doi:10.1002/dvdy.21176
Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE (2010) Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329(5990):444–448. doi:10.1126/science.1190485
Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, ten Hoeve J, Shuai K, Sun YE (2005) DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development 132(15):3345–3356. doi:10.1242/dev.01912
Jones PA, Baylin SB (2007) The epigenomics of cancer. Cell 128(4):683–692. doi:10.1016/j.cell.2007.01.029
Esteller M (2008) Epigenetics in cancer. N Engl J Med 358(11):1148–1159. doi:10.1056/NEJMra072067
Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O’Dorisio MS, Held WA, Cavenee WK, Plass C (2000) Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet 24(2):132–138. doi:10.1038/72785
Robertson KD (2005) DNA methylation and human disease. Nat Rev Genet 6(8):597–610. doi:10.1038/nrg1655
Urdinguio RG, Sanchez-Mut JV, Esteller M (2009) Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol 8(11):1056–1072. doi:10.1016/S1474-4422(09)70262-5
Sproul D, Meehan RR (2013) Genomic insights into cancer-associated aberrant CpG island hypermethylation. Brief Funct Genomics 12(3):174–190. doi:10.1093/bfgp/els063
Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, Hojo K, Yamanishi H, Karpf AR, Wallace DC, Simon M, Lander C, Boardman LA, Cunningham JM, Smith GE, Litchy WJ, Boes B, Atkinson EJ, Middha S, BD PJ, Parisi JE, Mer G, Smith DI, Dyck PJ (2011) Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet 43(6):595–600. doi:10.1038/ng.830
Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC (2004) Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep 5(12):1181–1186. doi:10.1038/sj.embor.7400295
Winkelmann J, Lin L, Schormair B, Kornum BR, Faraco J, Plazzi G, Melberg A, Cornelio F, Urban AE, Pizza F, Poli F, Grubert F, Wieland T, Graf E, Hallmayer J, Strom TM, Mignot E (2012) Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet 21(10):2205–2210. doi:10.1093/hmg/dds035
Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E (1999) Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402(6758):187–191. doi:10.1038/46052
Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM (1999) The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA 96(25):14412–14417
Jin B, Tao Q, Peng J, Soo HM, Wu W, Ying J, Fields CR, Delmas AL, Liu X, Qiu J, Robertson KD (2008) DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet 17(5):690–709. doi:10.1093/hmg/ddm341
de Greef JC, Wang J, Balog J, den Dunnen JT, Frants RR, Straasheijm KR, Aytekin C, van der Burg M, Duprez L, Ferster A, Gennery AR, Gimelli G, Reisli I, Schuetz C, Schulz A, Smeets DF, Sznajer Y, Wijmenga C, van Eggermond MC, Van Ostaijen-Ten Dam MM, Lankester AC, van Tol MJ, van den Elsen PJ, Weemaes CM, van der Maarel SM (2011) Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am J Hum Genet 88(6):796–804. doi:10.1016/j.ajhg.2011.04.018
Chouery E, Abou-Ghoch J, Corbani S, El Ali N, Korban R, Salem N, Castro C, Klayme S, Azoury-Abou Rjeily M, Khoury-Matar R, Debo G, Germanos-Haddad M, Delague V, Lefranc G, Megarbane A (2012) A novel deletion in ZBTB24 in a Lebanese family with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Clin Genet 82(5):489–493. doi:10.1111/j.1399-0004.2011.01783.x
Hou HA, Kuo YY, Liu CY, Chou WC, Lee MC, Chen CY, Lin LI, Tseng MH, Huang CF, Chiang YC, Lee FY, Liu MC, Liu CW, Tang JL, Yao M, Huang SY, Ko BS, Hsu SC, Wu SJ, Tsay W, Chen YC, Tien HF (2012) DNMT3A mutations in acute myeloid leukemia: stability during disease evolution and clinical implications. Blood 119(2):559–568. doi:10.1182/blood-2011-07-369934
Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK (2010) DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363(25):2424–2433. doi:10.1056/NEJMoa1005143
Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, Lein ES, Eadie BD, Willhoite AR, Muotri AR, Summers RG, Chun J, Lee KF, Gage FH (2003) Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci USA 100(11):6777–6782. doi:10.1073/pnas.1131928100
Li X, Barkho BZ, Luo Y, Smrt RD, Santistevan NJ, Liu C, Kuwabara T, Gage FH, Zhao X (2008) Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. J Biol Chem 283(41):27644–27652. doi:10.1074/jbc.M804899200
Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A (1992) Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69(6):905–914
Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY (2002) Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet 11(2):115–124
Chahrour M, Zoghbi HY (2007) The story of Rett syndrome: from clinic to neurobiology. Neuron 56(3):422–437. doi:10.1016/j.neuron.2007.10.001
Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP (2010) Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell 37(4):457–468. doi:10.1016/j.molcel.2010.01.030
Boyes J, Bird A (1991) DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64(6):1123–1134
Nan X, Campoy FJ, Bird A (1997) MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88(4):471–481
Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY (2009) Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet 18(13):2431–2442. doi:10.1093/hmg/ddp181
Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T (2005) Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet 37(1):31–40. doi:10.1038/ng1491
Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY (2005) Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci USA 102(49):17551–17558. doi:10.1073/pnas.0507856102
Adler DA, Quaderi NA, Brown SD, Chapman VM, Moore J, Tate P, Disteche CM (1995) The X-linked methylated DNA binding protein, Mecp2, is subject to X inactivation in the mouse. Mamm Genome 6(8):491–492
Schule B, Armstrong DD, Vogel H, Oviedo A, Francke U (2008) Severe congenital encephalopathy caused by MECP2 null mutations in males: central hypoxia and reduced neuronal dendritic structure. Clin Genet 74(2):116–126. doi:10.1111/j.1399-0004.2008.01005.x
Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23(2):185–188. doi:10.1038/13810
Ramocki MB, Peters SU, Tavyev YJ, Zhang F, Carvalho CM, Schaaf CP, Richman R, Fang P, Glaze DG, Lupski JR, Zoghbi HY (2009) Autism and other neuropsychiatric symptoms are prevalent in individuals with MeCP2 duplication syndrome. Ann Neurol 66(6):771–782. doi:10.1002/ana.21715
Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JA, Weitz CJ, Greenberg ME (2006) Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52(2):255–269. doi:10.1016/j.neuron.2006.09.037
Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, Wetsel WC, West AE, Greenberg ME (2011) Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron 72(1):72–85. doi:10.1016/j.neuron.2011.08.022
Guy J, Gan J, Selfridge J, Cobb S, Bird A (2007) Reversal of neurological defects in a mouse model of Rett syndrome. Science 315(5815):1143–1147. doi:10.1126/science.1138389
Giacometti E, Luikenhuis S, Beard C, Jaenisch R (2007) Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proc Natl Acad Sci USA 104(6):1931–1936. doi:10.1073/pnas.0610593104
Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG (2011) Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147(7):1498–1510. doi:10.1016/j.cell.2011.11.054
Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, Vermeulen M (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152(5):1146–1159. doi:10.1016/j.cell.2013.02.004
Muotri AR, Gage FH (2006) Generation of neuronal variability and complexity. Nature 441(7097):1087–1093. doi:10.1038/nature04959
Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH (2005) Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435(7044):903–910. doi:10.1038/nature03663
Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH (2009) L1 retrotransposition in human neural progenitor cells. Nature 460(7259):1127–1131. doi:10.1038/nature08248
Gatchel JR, Zoghbi HY (2005) Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet 6(10):743–755. doi:10.1038/nrg1691
La Spada AR, Taylor JP (2010) Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet 11(4):247–258. doi:10.1038/nrg2748
Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST (2007) Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron 55(4):556–564. doi:10.1016/j.neuron.2007.07.020
Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J (2007) RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron 55(4):565–571. doi:10.1016/j.neuron.2007.07.021
Ng CW, Yildirim F, Yap YS, Dalin S, Matthews BJ, Velez PJ, Labadorf A, Housman DE, Fraenkel E (2013) Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc Natl Acad Sci USA 110(6):2354–2359. doi:10.1073/pnas.1221292110
Cleary JD, Pearson CE (2003) The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet Genome Res 100(1–4):25–55. doi:72837
Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP et al (1991) Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65(5):905–914
Jin P, Alisch RS, Warren ST (2004) RNA and microRNAs in fragile X mental retardation. Nat Cell Biol 6(11):1048–1053. doi:10.1038/ncb1104-1048
Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas MF, Mandel JL (1991) Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science 252(5009):1097–1102
Williams K, Christensen J, Helin K (2012) DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep 13(1):28–35. doi:10.1038/embor.2011.233
Tan L, Shi YG (2012) Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 139(11):1895–1902. doi:10.1242/dev.070771
Branco MR, Ficz G, Reik W (2012) Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet 13(1):7–13. doi:10.1038/nrg3080
Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I (2011) TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell 9(3):193–204. doi:10.1016/j.stem.2011.08.007
Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T (2011) The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem 50(31):7008–7012. doi:10.1002/anie.201103899
Guo JU, Su Y, Zhong C, Ming GL, Song H (2011) Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145(3):423–434. doi:10.1016/j.cell.2011.03.022
Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C (2012) Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol 8(4):328–330. doi:10.1038/nchembio.914
Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X (2012) Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 40(11):4841–4849. doi:10.1093/nar/gks155
Inoue A, Shen L, Dai Q, He C, Zhang Y (2011) Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res 21(12):1670–1676. doi:10.1038/cr.2011.189
Mayer W, Niveleau A, Walter J, Fundele R, Haaf T (2000) Demethylation of the zygotic paternal genome. Nature 403(6769):501–502. doi:10.1038/35000654
Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J (2000) Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10(8):475–478
Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL (2011) The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477(7366):606–610. doi:10.1038/nature10443
Vincent JJ, Huang Y, Chen PY, Feng S, Calvopina JH, Nee K, Lee SA, Le T, Yoon AJ, Faull K, Fan G, Rao A, Jacobsen SE, Pellegrini M, Clark AT (2013) Stage-Specific Roles for Tet1 and Tet2 in DNA Demethylation in Primordial Germ Cells. Cell Stem Cell. doi:10.1016/j.stem.2013.01.016
Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA (2012) Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. doi:10.1126/science.1229277
Kress C, Thomassin H, Grange T (2001) Local DNA demethylation in vertebrates: how could it be performed and targeted? FEBS Lett 494(3):135–140
Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C (2007) Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445(7128):671–675. doi:10.1038/nature05515
Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H (2009) Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323(5917):1074–1077. doi:10.1126/science.1166859
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19(1):17–30. doi:10.1016/j.ccr.2010.12.014
Losman JA, Looper R, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley G, Root D, Ebert BL, Kaelin WG Jr (2013) (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. doi:10.1126/science.1231677
Liang DC, Liu HC, Yang CP, Jaing TH, Hung IJ, Yeh TC, Chen SH, Hou JY, Huang YJ, Shih YS, Huang YH, Lin TH, Shih LY (2013) Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2 and DNMT3A. Blood. doi:10.1182/blood-2012-06-436782
Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, Lee CW, Hu D, Lian BQ, Kleffel S, Yang Y, Neiswender J, Khorasani AJ, Fang R, Lezcano C, Duncan LM, Scolyer RA, Thompson JF, Kakavand H, Houvras Y, Zon LI, Mihm MC Jr, Kaiser UB, Schatton T, Woda BA, Murphy GF, Shi YG (2012) Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150(6):1135–1146. doi:10.1016/j.cell.2012.07.033
Gorovets D, Kannan K, Shen R, Kastenhuber ER, Islamdoust N, Campos C, Pentsova E, Heguy A, Jhanwar SC, Mellinghoff IK, Chan TA, Huse JT (2012) IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin Cancer Res 18(9):2490–2501. doi:10.1158/1078-0432.CCR-11-2977
Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, Pasini D (2013) Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. doi:10.1016/j.molcel.2012.12.019
Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. doi:10.1038/emboj.2012.357
Chen Q, Chen Y, Bian C, Fujiki R, Yu X (2012) TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. doi:10.1038/nature11742
Francisco H, Kollins K, Varghis N, Vocadlo D, Vosseller K, Gallo G (2009) O-GLcNAc post-translational modifications regulate the entry of neurons into an axon branching program. Dev Neurobiol 69(2–3):162–173. doi:10.1002/dneu.20695
Liu Y, Li X, Yu Y, Shi J, Liang Z, Run X, Li Y, Dai CL, Grundke-Iqbal I, Iqbal K, Liu F, Gong CX (2012) Developmental regulation of protein O-GlcNAcylation, O-GlcNAc transferase, and O-GlcNAcase in mammalian brain. PLoS ONE 7(8):e43724. doi:10.1371/journal.pone.0043724
Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q (2013) Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. doi:10.1016/j.celrep.2013.01.011
Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, Diao J, Wu F, He HH, Cui Q, Clark E, Ma C, Barbara A, Veenstra GJ, Xu G, Kaiser UB, Liu XS, Sugrue SP, He X, Min J, Kato Y, Shi YG (2012) Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus Eye and neural development. Cell 151(6):1200–1213. doi:10.1016/j.cell.2012.11.014
Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, Das S, Levasseur DN, Li Z, Xu M, Reik W, Silva JC, Wang J (2013) NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. doi:10.1038/nature11925
Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, Levine RL, Nik S, Chen EI, Abeliovich A (2012) Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488(7413):652–655. doi:10.1038/nature11333
Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K (2011) TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473(7347):343–348. doi:10.1038/nature10066
Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, Min J, Nicholson T, Chen T, Xu G, Shi Y, Zhang K, Shi YG (2011) Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42(4):451–464. doi:10.1016/j.molcel.2011.04.005
Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A (2011) Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8(2):200–213. doi:10.1016/j.stem.2011.01.008
Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, Jaenisch R (2011) Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9(2):166–175. doi:10.1016/j.stem.2011.07.010
Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, Faull KF, Lyko F, Jaenisch R (2013) Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. doi:10.1016/j.devcel.2012.12.015
Wang F, Yang Y, Lin X, Wang JQ, Wu YS, Xie W, Wang D, Zhu S, Liao YQ, Sun Q, Yang YG, Guo C, Han C, Tang TS (2013) Genome-wide loss of 5-hmC is a novel epigenetic feature of huntington’s disease. Hum Mol Genet. doi:10.1093/hmg/ddt214
Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, Gao J, Liu P, Li L, Xu GL, Jin P, He C (2013) Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. doi:10.1016/j.cell.2013.04.001
Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, Zhang K, Zhang Y (2013) Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. doi:10.1016/j.cell.2013.04.002
Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A (2010) Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468(7325):839–843. doi:10.1038/nature09586
Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD, Melnick A, Godley LA, Aifantis I, Levine RL (2011) Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20(1):11–24. doi:10.1016/j.ccr.2011.06.001
Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A (2011) Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA 108(35):14566–14571. doi:10.1073/pnas.1112317108
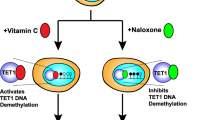
Minor EA, Court BL, Young JI, Wang G (2013) Ascorbate induces Ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem. doi:10.1074/jbc.C113.464800
Acknowledgments
The authors would like to thank C. Strauss for critical reading of the manuscript. P.J. is supported by NIH grants (NS079625/NS051630/HD073162/AG025688 to P.J.), and the Simons Foundation Autism Research Initiative (P.J.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yao, B., Jin, P. Cytosine modifications in neurodevelopment and diseases. Cell. Mol. Life Sci. 71, 405–418 (2014). https://doi.org/10.1007/s00018-013-1433-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-013-1433-y