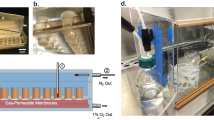
Abstract
The importance of the gut microbiota in human health and disease progression makes it a target for research in both the biomedical and nutritional fields. To date, a number of in vitro systems have been designed to recapitulate the gut microbiota of the colon ranging in complexity from the application of a single vessel to cultivate the community in its entirety, to multi-stage systems that mimic the distinct regional microbial communities that reside longitudinally through the colon. While these disparate types of in vitro designs have been employed previously, information regarding similarities and differences between the communities that develop within was less defined. Here, a comparative analysis of the population dynamics and functional production of short-chain fatty acids (SCFAs) was performed using the gut microbiota of the same donor cultured using a single vessel and a 3-stage colon system. The results found that the single vessel communities maintained alpha diversity at a level comparable to the distal regions of the 3-stage colon system. Yet, there was a marked difference in the type and abundance of taxa, particularly between families Enterobacteriaceae, Bacteroidaceae, Synergistaceae, and Fusobacteriaceae. Functionally, the single vessel community produced significantly less SCFAs compared to the 3-stage colon system. These results provide valuable information on how culturing technique effects gut microbial composition and function, which may impact studies relying on the application of an in vitro strategy. This data can be used to justify experimental strategy and provides insight on the application of a simplified versus complex study design.
Key points
• A mature gut microbiota community can be developed in vitro using different methods.
• Beta diversity metrics are affected by the in vitro culturing method applied.
• The type and amount of short-chain fatty acids differed between culturing methods.







Similar content being viewed by others
Availability of data and material
All data will be made available upon request.
Code availability
Not applicable.
References
Allen-Vercoe E, Strauss J, Chadee K (2011) Fusobacterium nucleatum: an emerging gut pathogen? Gut Microbes 2:294–298
Aura AM, Maukonen J (2015) One compartment fermentation model. In: Verhoeckx K, Cotter P, López-Expósito I, Kleiveland C, Lea T, Mackie A, Requena T, Swiatecka D, Wichers H (eds) The impact of food bioactives on health: in vitro and ex vivo models. Springer, Cham, pp 281–292
Bhattacharya T, Ghosh T, Mande S (2015) Global profiling of carbohydrate active enzymes in human gut microbiome. PLoS One 10:e0142038
Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J (2018) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, MGI L, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, Mc Donald D, LJ MI, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, vander Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 47:852–857
Brennan C, Garrett W (2019) Fusbacterium nucleatum-symbiont, opportunist and oncobacterium. Nat Rev Microbiol 17:156–166
Brüssow H (2020) Problems with the concept of gut microbiota dysbiosis. Microb Biotechnol 13:423–434
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583
Clarke G, Stilling R, Kennedy P, Stanton C, Cryan J, Dinan T (2014) Minireview: gut microbiota: the neglected endocrine organ. Mol Endocrinol 28:1221–1238
Cockburn D, Koropatkin N (2016) Polysaccharide degradation by the intestinal microbiota and it’s influence on human health and disease. J Mol Biol 428:3230–3252
Cummings J, Macfarlane G (1991) The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol 70:443–459
Dey P (2019) Gut microbiota in phytopharmacology: a comprehensive overview of concepts, reciprocal interactions, biotransformations and mode of actions. Pharmacol Res 147:104367
Donaldson G, Lee S, Mazmanian S (2015) Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14:20–32
Fallingborg J (1999) Intraluminal pH of the human gastrointestinal tract. Dan Med Bull 46:183–196
Firrman J, Liu L, Tanes C, Friedman E, Bittinger K, Daniel S, Van den Abbeele P, Evans B (2019a) Metabolic analysis of regionally distinct gut microbial communities using an in vitro platform. J Agric Food Chem 68:13056–13067
Firrman J, Liu L, Van den Abbeele P, Tanes C, Bittinger K, Tomasula P (2019b) Applying advanced in vitro culturing technology to study the human gut microbiota. J Vis Exp 15:144
Franzosa E, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser H, Reinker S, Vatanen T, Hall A, Mallick H, McIver L, Sauk J, Wilson R, Stevens B, Scott J, Pierce K, Deik A, Bullock K, Imhann F, Porter J, Zhernakova A, Fu J, Weersma R, Wijimenga C, Clish C, Vlamakis H, Huttenhower C, Xavier R (2019) Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 4:293–305
Gibson G, Macfarlane S, Macfarlane G (1993) Degradative activities of gut anaerobes studied in a three-stage continuous culture model of the colon. Clin Infect Dis 16:S420–S421
Guzman-Rodriguez M, McDonald J, Hyde R, Allen-Vercoe E, Claud E, Sheth P, Petrof E (2018) Using bioreactors to study the effects of drugs on the human microbiota. Methods 149:31–41
IIhan Z, Marcus A, Kang D, Rittmann B, Krajmalnik-Brown R (2017) pH-mediated microbial and metabolic interactions in fecal enrichment cultures. MSphere 2:e0047–e0014
James KR, Gomes T, Elmentaite R, Kumar N, Gulliver EL, King HW, Stares MD, Bareham BR, Ferdinand JR, Petrova VN, Polanski K, Forster SC, Jarvis LB, Suchanek O, Howlett S, James LK, Jones JL, Meyer KB, Clatworthy MR, Saeb-Parsy K, Lawley TD, Teichmann SA (2020) Distinct microbial and immune niches of the human colon. Nat Immunol 21:343–353
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kaoutari A, Armougom F, Gordon J, Raoult D, Henrissat B (2013) The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504
Liu L, Firrman J, Tanes C, Bittinger K, Thomas-Gahring A, Wu G, Van den Abbeele P, Tomasula P (2018) Establishing a mucosal gut microbial community in vitro using an artificial simulator. PLoS One 13:e0197692
Luca M, Di Mauro M, Di Mauro M, Luca A (2019) Gut microbiota in Alzheimer’s disease, depression, and type 2 diabetes mellitus: the role of oxidative stress. Oxidative Med Cell Longev 2019:4730539
Lynch S, Pedersen O (2016) The human intestinal microbiome in health and disease. N Engl J Med 375:2369–2379
Macfarlane G, Gibson G, Cummings J (1992) Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol 72:57–64
Macfarlane G, Macfarlane S, Gibson G (1998) Validation of a three-stage compound continuous culture system for investigating the effects of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol 35:180–187
Mahalak KK, Firrman J, Tomasula PM, Nuñez A, Lee JJ, Bittinger K, Rinaldi W, Liu LS (2020) Impact of steviol glycosides and erythritol on the human and Cebus apella gut microbiome. J Agric Food Chem 68:13093–13101
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618
McDonald J, Fuentes S, Schroeter K, Heikamp-deJong I, Khursigara C, de Vos W, Allen Vercoe E (2015) Simulating distal gut mucosal and luminal communities using packed-column biofilm reactors and an in vitro chemostat model. J Microbiol Methods 108:26–44
McDonald J, Schroeter K, Fuentes S, Heikamp-deJong I, Khursigara C, de Vos W, Allen-Vercoe E (2013) Evaluation of microbial community reproducibility, stability and composition in a human distal gut chemostat model. J Microbiol Methods 95:167–174
Messer J, Liechty E, Vogel O, Chang E (2017) Evolutionary and ecological forces that shape the bacterial communities of the human gut. Mucosal Immunol 10:567–579
Molly K, De Smet I, Nollet L, Van de Woestyne M, Verstraete W (1996) Effect of Lactobacilli on the ecology of the gastro-intestinal microbiota cultured in the SHIME reactor. Microb Ecol Health Dis 9:79–89
Molly K, Van de Woestyne M, De Smet I, Verstraete W (1994) Validation of the simulator of the human intestinal microbial ecosystem (SHIME) reactor using microorganism-associated activities. Microb Ecol Health Dis 7:191–200
Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650
Schoeler M, Caesar R (2019) Dietary lipids, gut microbiota and lipid metabolism. Rev Endocr Metab Diosord 20:461–472
Strandwitz P (2018) Neurotransmitter modulation by the gut microbiota. Brain Res 1693:128–133
Takagi R, Sasaki K, Sasaki D, Fukuda I, Tanaka K, Yoshida K, Kondo A, Osawa R (2016) A single-batch fermentation system to simulate human colonic microbiota for high-throughput evaluation of prebiotics. PLoS One 11:e0160533
Tang WH, Kitai T, Hazen SL (2017) Gut microbiota in cardiovascular health and disease. Circ Res 120:1183–1196
Tsitko I, Wiik-Miettinen F, Mattila O, Rosa-Sibakov N, Seppanen-Laakso T, Maukonen J, Nordlund E, Saarela M (2019) A small in vitro fermentation model for screening the gut microbiota effects of different fiber preparations. Int J Mol Sci 20:1925
Van de Wiele T, Van den Abbeele P, Ossieur W, Possemiers S, Marzorati M (2015) The simulator of the human intestinal microbial ecosystem (SHIME®). In: Verhoeckx K, Cotter P, López-Expósito I, Kleiveland C, Lea T, Mackie A, Requena T, Swiatecka D, Wichers H (eds) The impact of food bioactives on health: in vitro and ex vivo models. Springer, Cham, pp 305–317
Van den Abbeele P, Grootaert C, Marzorati M, Possemiers S, Verstraete W, Gerard P, Rabot S, Bruneau A, El Aidy S, Derrien M, Zoetendal E, Kleerebezem M, Smidt H, Van de Wiele T (2010) Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl Environ Microbiol 76:5237–5246
Venema K (2015) The TNO in vitro model of the colon (TIM-2). In: Verhoeckx K, Cotter P, López-Expósito I, Kleiveland C, Lea T, Mackie A, Requena T, Swiatecka D, Wichers H (eds) The impact of food bioactives on health: in vitro and ex vivo models. Springer, Cham, pp 293–304
Vrancken G, Gregory A, Huys G, Faust K, Raes J (2019) Synthetic ecology of the human gut microbiota. Nat Rev Microbiol 17:754–763
White B, Lamed R, Bayer E, Flint H (2014) Biomass utilization by gut microbiomes. Annu Rev Microbiol 68:279–296
Wu G, Lewis J, Hoffmann C, Chen Y, Knight R, Bittinger K, Hwang J, Chen J, Berkowsky R, Nessel L, Li H, Bushman F (2010) Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol 10:206
Yadav M, Verma M, Chauhan N (2018) A review of metabolic potential of human gut microbiome in human nutrition. Arch Microbiol 200:203–217
Yoav B, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B 57:289–300
Funding
This work was supported by the in-house Project 8072-41000-102-00D, “In Vitro Human Intestinal Microbial Ecosystem: Effects of Diet.”
Author information
Authors and Affiliations
Contributions
JF, KM, and LSL designed and performed the experiments; LM and HZ performed Next Gen DNA sequencing; CT and KB performed the bioinformatical and statistical analyses; JB performed metabolomics; PVA assisted in experimental design; JF, KM, LSL, LM, HZ, CT, KB, JB, and PVA contributed to writing and revising the manuscript and interpreting the results; and JF was responsible for compiling the data and coordinating the research efforts.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 646 kb)
Rights and permissions
About this article
Cite this article
Firrman, J., Liu, L., Mahalak, K. et al. Comparative analysis of the gut microbiota cultured in vitro using a single colon versus a 3-stage colon experimental design. Appl Microbiol Biotechnol 105, 3353–3367 (2021). https://doi.org/10.1007/s00253-021-11241-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11241-x