Abstract.
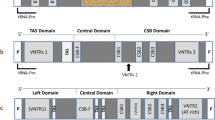
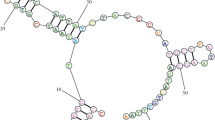
The entire mitochondrial DNA control region (mtDNA D-loop) was sequenced in the seven extant species of Alectoris partridges. The D-loop length is very conserved (1155 ± 2 nucleotides), and substitution rates are lower than for the mitochondrial cytochrome b gene of the same species, on average. Comparative analyses suggest that these D-loops can be divided into three domains, corresponding to the highly variable peripheral domains I and III and to the central conserved domain II of vertebrates (Baker and Marshall 1997). Nevertheless, the first 161 nucleotides of domain I of the Alectoris, immediately flanking the tRNAGlu, evolve at an unusually low rate and show motifs similar to the mammalian extended termination-associated sequences [ETAS1 and ETAS2 (Sbisà et al. 1997)], which can form stable secondary structures. The second part of domain I contains a hypervariable region with two divergent copies of a tandemly repeated sequence described previously in other species of anseriforms and galliforms (Quinn and Wilson 1993; Fumihito et al. 1995). Some of the conserved sequence blocks of mammals can be mapped in the central domain of Alectoris. Domain III is highly variable and has sequences similar to mammalian CSB1. The bidirectional transcription promoter HSP/LSP box of the chicken is partially conserved among the Alectoris. This structural organization can be found in the anseriform and galliform species studied so far, suggesting that strong functional constraints might have controlled the evolution of the D-loop since the origin of Galloanserae. Their conserved organization and slow molecular evolution make D-loops of galliforms appropriate for phylogenetic studies, although homoplasy can be be generated at a few hypervariable sites and at some sites which probably have mutated by strand slippage during DNA replication. Phylogenetic analyses of D-loops of Alectoris are concordant with previously published cytochrome b and allozyme phylogenies (Randi 1996). Alectoris is monophyletic and includes three major clades: (1) basal barbara and melanocephala; (2) intermediate rufa and graeca; and (3) recent philbyi, magna, and chukar. Comparative description of the organization and substitution patterns of the mitochondrial control region can aid in mapping hypervariable sites and avoid some sources of homoplasy in data sets which are to be used in phylogenetic analyses.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 18 August 1997 / Accepted: 20 February 1998
Rights and permissions
About this article
Cite this article
Randi, E., Lucchini, V. Organization and Evolution of the Mitochondrial DNA Control Region in the Avian Genus Alectoris . J Mol Evol 47, 449–462 (1998). https://doi.org/10.1007/PL00006402
Issue Date:
DOI: https://doi.org/10.1007/PL00006402