Abstract
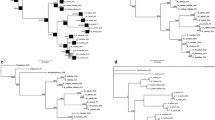
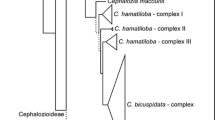
We sequenced the nuclear ribosomal internal transcribed spacer (ITS) regions to determined the phylogenetic relationships of the severalGoodyera species in Korea and to measure the extent of their differentiation region. ITS 1 was 238 to 239 bp long while ITS 2 was 258 to 259 bp. The 5.8S coding region was 156 bp long. Sequence divergences among species, calculated by Kimura’s two- parameter method, ranged from 0.0 to 5.4%. The most parsimonious tree, with a consistency index of 0.935 and a retention index of 0.937, was produced with 337 steps. Our ITS sequence results demonstrate the monophyly of KoreanGoodyera and support previous morphological, geographical, and RAPD data analyses.
Similar content being viewed by others
Literature Cited
Ackerman JD (1975) Reproductive biology ofGoodyera oblongifolia (Orchidaceae). Madrono23: 191–198
Baldwin BG (1992) Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: An example from Compositae. Molec Phylog Evol1: 3–16
Baldwin GB (1993) Molecular phylogenetics of Calycadenia (Compositae) based on ITS sequences of nuclear ribosomal DNA: Chromosomal and morphological evolutionary reexamined. Amer J Bot80: 222–238
Baldwin GB, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA: A valuable source of evidence on angiosperm phylogeny. Ann Missouri Bot Gard82: 247–277
Cho HJ, Kim St, Suh YB, Park CW (1996) ITS sequences of someAcer species and phylogenetic implication. Kor J Plant Tax26: 271–291
Choi B-H, Kim J-H (1997) ITS sequences and speciation on Far EasternIndigofera (Leguminosae). J Plant Res110: 339–346
Choi HK, Kim CK, Shin HC (2000) Molecular reexamination of Korean Umbelliferae based on internal transcribed spacer sequences of rDNA:Ligusticum tenuissimum (Nakai) Kitagawa andLibanotis coreana (Wolff) Kitagawa. J Plant Biol43: 136–142
Choi HK, Kim HJ, Shin HC, Kim YD (1996) Phylogeny and ribosomal DNA variations ofBupleurum (Umbelliferae). Kor J Plant Tax26: 219–233
Douzery EJ, Pridgeon AM, Kores P, Under HP, Kurzweil H, Chase MW (1999) Molecular phylogenetics of Diseae (Orchidaceae): A contribution from nuclear ribosomal ITS sequences. Amer J Bot86: 887
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull19: 11–15
Eftimiu-Heim P (1941) Recherches surles noyau des Orchidees. Paris Le Botaniste31: 65–111
Erdogan V, Mehlenbacher SA (2000) Phylogenetic relationships ofCorylus species (Betulaceae) based on nuclear ribosomal DNA ITS region and chloroplast matK gene sequences. Syst Bot4: 727–737
Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution39: 783–791
Femald ML (1950) Cray’s Manual of Botany. Amer Book Co, pp 480–481
GenBank (2002) Nucl Acids Res30: 17–20
Goldberg A (1986) Classification, evolution and phylogeny of the families of dicotyledons. Smithsonian Institution, Washington DC
Ikuse M (1956) Pollen Grains of Japan. Hirokawa Pub Co, Tokyo
Kallunki JA (1976) Population studies in Goodyera (Orchidaceae) with emphasis on the hybrid origin of C.tesselata. Brittonia28: 53–75
Kim K-J, Jansen RK (1994) Comparisons of phylogenetic hypotheses among different data sets in dwarf dandelions (Krigia): Additional information from internal transcribed spacer sequences of nuclear ribosomal DNA. Pl Syst Evol190: 157–185
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparable studies of nucleotide sequences. J Molec Evol16: 111–120
Kitagawa M (1939) Lineamenta Florae Manshuricae. Inst Sci Res Manchoukuo2: 487
Kitamura SG, Murata, Koyama T (1980) Colored Illustrations of Herbaceous Plants of Japan (Monocotyledoneae). Hoikusa Pub Co, LTD, Tokyo
Konta F, Tsuji M (1982) The types of pollen tetrads and their formations observed in some species in the Orchidaceae in Japan. Acta Phytotax Geobot33: 206–217
Lee TB (1980) Illustrated Flora of Korea. Hyangmoonsa Seoul, pp 243–244
Li HL (1978) Flora of Taiwan. Vol 5. Epoch Pub Co, Taipei, Taiwan pp 1006–1019
Love A (1954) Cytotaxonomical evaluation of corresponding taxa. Vegetatio5–6: 212–224
Love A, Love D (1981) In chromosome number reports LXX III. Taxon30: 845–851
Mutsuura O, Nakahira R (1960) Chromosome numbers of the family Orchidaceae in Japan (3). Sci Rep Kyoto Pre Univ 3A, Ser: 83–88
Ohwi J (1984) Flora of Japan. Ed 2, Smithsonian Inst Washington DC, pp 339–340
Richardson MM (1935) The chromosomes of some British orichids. Proc Univ Durham Philos Soc9: 135–140.
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol4: 406–425
Satake YJ, Tominari T (1982) Wild Flowers of Japan. Monoco.[1]. Heibonsha, Tokyo pp 211–213
Sun BY, Kim TJ, Kim ST (2000) Systematics ofOstericum (Apiaceae) in Korea. Kor J Plant Tax30: 93–104
Swofford DL (2001) PAUP - Phylogenetic analysis using parsimony (and other methods). Ver. 4.06b. Academic Press, Sunderland, MA
Tae KH, In DS, Bae HY, Ko SC (1999) Relationship of the KoreanCoodyera (Orchidaceae) using random amplified polymorphic DNAs analysis. Kor J Plant Tax29: 169–181
Tae KH, Ko SC (1999) Interspecific relationship of the genusGoodyera in Korea using cluster analysis. Kor J Plant Tax29: 63–73
Tae KH, Lee EH, Ko SC (1997) A systematic study of the genus Goodyera in Korea by morphological and cytological characters. Kor J Plant Tax27: 89–116
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: Flexible strategies for multiple sequence aligment aided by quality analysis tools. Nucl Acids Res25: 4876–4882
Ueno J (1976) The fine structure of pollen surface II Orchidaceae. Rep Fac Sci Shizuoka Univ11: 87–109
Vij SR Gupta GC (1975) Cytological investigations into W. Himalayan Orchidaceae I. Chromosome numbers and karyotypes of taxa from Kashmir. Cytologia40: 613–621
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,In M Innis, D Gelgand, J Snisky, T White, eds, PCR Protocols: A Guide to Methods and Amplification, Academic Press, San Diego, pp 315–322
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shin, KS., Shin, Y.K., Kim, JH. et al. Phylogeny of the genusGoodyera (Orchidaceae; Cranichideae) in Korea based on nuclear ribosomal DNA ITS region sequences. J. Plant Biol. 45, 182–187 (2002). https://doi.org/10.1007/BF03030312
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03030312