Abstract
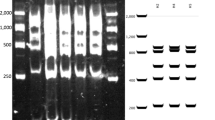
Molecular markers for alder,Alnus firma Sieb. et Zucc, have not been studied extensively. Here, we used amplified fragment length polymorphism (AFLP) to investigate genetic relationships among 15 natural populations. EcoRI-ACG + Msel-CTG combinations revealed the highest polymorphism (62.2%). A total of 171 DNA fragments were identified. On average, 58.1% of the AFLP markers that were generated using four primer pairs were polymorphic. Diversity was insignificant among the populations. The combination of a wind-pollinated, outcrossing breeding system along with large population sizes, and the ability to regenerate by stump sprouting may explain the high level of genetic diversity within this species. The majority (98%) of the genetic variance resided within populations. The average number of individuals that were exchanged between populations per generation was very high (N em = 12.3). Gene dispersal in alder is apparently by seed dispersalvia water and human activity as well as through pollen. Five individuals per population were claded in the same cluster.
Similar content being viewed by others
Literature Cited
Aggarwal RK, Brar DS, Nandi S, Huang N, Khush GS (1999) Phylogenetic relationships amongOriza species revealed by AFLP markers. Theor Appl Genet98: 1320–1328
An K, An G (2000) Overriding Photoperiod Sensitivity of Flowering Time by Constitutive Expression of a MADS Box Gene. J Plant Biol43: 28–32
Bousquet J, Cheliak WM, Lalonde M (1987a) Allozyme variability in natural populations of green alder(Alnus crispa) in Quebec. Genome29: 345–352
Bousquet J, Cheliak WM, Lalonde M (1987b) Genetic differentiation among 22 mature populations of green alder (A/nuscrispa) in Central Quebec. Can J For Res17: 219–227
Bousquet J, Cheliak WM, Lalonde M (1988) Allozyme variation within and among mature populations of speckled alder (A/nusrugosa) and relationships with green alder (Acrispa). Am J Bot75: 1678–1686
Bowman KD, Hutcheson K, Odum EP, Shenton LR (1971) Comments on the distribution of indices of diversity. StatE col3: 315–359
Felsenstein J (1993) PHYLIP (Phylogeny Inference Package) version 3.5s. Distributed by the author. Department of Genetics, Univ Washington, Seattle
Furlow JJ (1979) The systematics of the American species ofAlnus (Betulaceae). Rhodora81: 1–121
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forests6: 95–124
Hongtrackul V, Huestis GS, Knapp SJ (1997) Amplified fragment length polymorphisms as a tool for DNA fingerprint of sunflower germplasm: genetic diversity among oilseed inbred lines. Theor Appl Genet95: 400–407
Huenneke LR (1985) Spatial distribution of genetic individuals in thickets ofAlnus incana ssp.rugosa, a clonal shrub. Am J Bot72: 152–158
Huenneke LR (1987) Demography of a clonal shrub,Alnus incana ssp.rugosa (Betulaceae). Am Midi Nat117: 43–55
Huh MK (1999) Genetic diversity and population structure of Korean Alder (A/nusjaponica: Betulaceae). Can J For Res29: 1311–1316
Huh MK, Huh HW (1999) Genetic diversity and population structure ofAlnus hirsuta (Betulaceae). J Plant Res112: 437–442
Karron JD (1987) A comparison of levels of genetic polymorphism in geographically restricted and widespread plant congenera. Evol Ecol1: 47–58
Kim S-R, An G (1996) Characterization of an Easter Lily Calmodulin cDNA Clone. J Plant Biol39: 9–13
King LM, Schaal BA (1989) Ribosomal DNA variation and distribution ofRudbeckia missouriensis. Evolution42: 1117–1119
Le Thierry diEnneequin M, Poupance B, Starr A (2000) Assessment of genetic relationships betweenSetaria ital-ica and its wild relativeS. viridis using AFLP markers. Theor Appl Genet100: 1061–1066
Maheswaran M, Subudhi PK, Nandi S, Xu JC, Parco A, Yang DC, Huang N (1997) Polymorphism, distribution, and segregation of AFLP markers in a doubled-haploid rice population. Theor Appl Genet94: 39–45
Maughan PJ, Maroof MAS, Buss GR, Huestis GM (1996) Amplified fragment length polymorphism (AFLP) in soybean: species diversity, inheritance, and near-isogenic line analysis. Theor Appl Genet93: 392–401
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA74: 5267–5273
Normand R Lalonde M (1986) The genetics ofFrankia: a review. Plant Soil90: 429–453
Paul SP, Wachira FN, Powell W, Waugh R (1997) Diversity and genetic differentiation among populations of Indian and Kenyan tea(Camellia sinensis (L.) O. Kuntze) revealed by AFLP markers. Theor Appl Genet94: 255–263
Powell W, Michele M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLR RAPD, AFLP and SSR (microsatellite) markers for germ-plasm analysis. Mol Breed2: 225–238
Purdy BG, Bayer RL, Macdonald SR (1994) Genetic variation, breeding system evolution, and conservation of the narrow sand dune endemicStellaria arenicola and the widespreadS. longipes (Caryophyllaceae). Am J Bot81: 904–911
Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol4: 406–425
Singh A, Negi MS, Rajagopal J, Bhatia S, Tomar UK, Srivas-tava PS, Lakshmikumaran M (1999) Assessment of genetic diversity inAzadirachta indica using AFLP markers. Theor Appl Genet99: 272–279
Smouse PE, Long JC, Sokal RR (1986) Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst Zool35: 627–632
Teulat B, Aldam C, Trehin R, Lebrun R Barker JHA, Arnold GM, Karp A, Baudouin L (2000) An analysis of genetic diversity in coconut(Cocos nucifera) populations from across the geographic range using sequence-tagged microsatellites (SSRs) and AFLPs. Theor Appl Genet100: 764–771
Vos P, Hogers R, Bleeker M, Reijans M, van Lee T, Hornes M, Freijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acid Res23: 4407–4414
Woodland DW (1991) Contemporary Plant Systematics. Prentice-Hall, Inc, Englewood Cliffs, NJ, pp 222
Wright S (1951) The genetic structure of populations. Ann Eugen15: 313–354
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Huh, M.K., Huh, H.W. Genetic diversity and phylogenetic relationships in alder,Alnus firma, revealed by AFLP. J. Plant Biol. 44, 33–40 (2001). https://doi.org/10.1007/BF03030274
Issue Date:
DOI: https://doi.org/10.1007/BF03030274