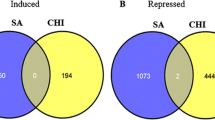
Abstract
Microarray analysis makes it possible to determine the relative expression of thousands of genes simultaneously. It has gained popularity at a rapid rate, but many caveats remain. In an effort to establish reliable microarray protocols for sweetpotato [Ipomoea batatas (L.) Lam.], we compared the effect of replication number and image analysis software with results obtained by quantitative rela-time PCR (Q-RT-PCR). Sweetpotato storage root development is the most economically important process in sweetpotato. In order to identify genes that may play a role in this process, RNA for microarray analysis was extracted from sweetpotato fibrous and storage roots. Four data sets, Spot4, Spot6, Finder4 and Finder6, were created using 4 or 6 replications, and the image analysis software of UCSF Spot or TIGR Spotfinder were used for spot detection and quantification. The ability of these methods to identify significant differential expression between treatments was investigated. The data sets with 6 replications were better at identifying genes with significant differential expression than the ones of 4 replications. Furthermore when using 6 replicates, UCSF Spot was superior to TIGR Spotfinder in identifying genes differentially expressed (18 out of 19) based on Q-RT-PCR. Our study shows the importance of proper replication number and image analysis for microarray studies.
Similar content being viewed by others
Abbreviations
- AGPase:
-
ADP-glucose pyrophosphorylase
- GAPDH:
-
glyceraldehyde-3-phosphate dehydrogenase
- limma:
-
Linear Models for Microarray Data
- Q-RT-PCR:
-
quantitative real-time PCR
- SuSy:
-
Sucrose Synthase
- TAIR:
-
The Arabidopsis Information Resource
- TIGR:
-
The Institute for Genomic Research
References
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, and Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29.
Bas A, Forsberg G, Hammarström S, and Hammarström M-L (2004) Utility of the house-keeping genes 18S rRNA, β-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol 59: 566–573.
Benjamini Y and Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300.
Brinker M, van Zyl L, Liu W, Craig D, Sederoff RR, Clapham DH, and von Arnold S (2004) Microarray analyses of gene expression during adventitious root development inPinus contorta. Plant Physiol 135: 1526–1539.
Churchill GA (2002) Fundamentals of experimental design for cDNA microarrays. Nat Genet suppl. 32: 490–495.
Doyle JJ and Doyle JL (1987) Isolation of plant material from fresh tissue. Focus 12: 13–15.
Drăghici S (2003) Data analysis tools for DNA microarrays. Chapman & Hall/CRC, London, UK.
Ewing B and Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194.
Food and Agricultural Organization (1993) 1992 Production year-book. Food and Agricultural Organization statistics series vol. 46, No. 112, Rome.
Gilliland LU, Kandasamy MK, Pawloski LC, and Meagher RB (2002) Both vegetative and reproductive actin isovariants complement the stunted root hair phenotype of the Arabidopsis act2-1 mutation. Plant Physiol 130: 2199–2209.
Glisin V, Crkvenjakov R, and Byus C (1974) Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry 13: 2633–2637.
Goldsmith ZG and Dhanasekaran N (2004) The microrevolution: applications and impacts of microarray technology on molecular biology and medicine (review). Int J Mol Med 13: 483–495.
Holm S (1979) A simple sequentially rejective Bonferroni test procedure. Scand J Stat 6: 65–70.
Huang X and Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9: 868–877.
Hussey PJ, Haas N, Hunsperger J, Larkin J, Snustad DP, and Silflow CD (1990) The b-tubulin gene family in Zea mays: two differentially expressed b-tubulin genes. Plant Mol Biol 15: 957–972.
Iskandar HM, Simpson RS, Casu RE, Bonnett GD, MaClean DJ, and Manners JM (2004) Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol Biol Rep 11: 325–337.
Jain AN, Tokuyasu TA, Snijders AM, Segraves R, Albertson DG, and Pinkel D (2002) Fully automatic quantification of microarray data. Genome Res 12: 325–332.
Joyce CM, Villemur R, Snustad DP, and Silflow CD (1992) Change in isotype expression along the developmental axis of seedling root. J Mol Biol 227: 97–107.
Kendziorski C, Irizarry RA, Chen K-S, Haag JD, and Gould MN (2005) On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci USA 102(12): 4252–4257.
Kim B-R, Nam H-Y, Kim S-U, Kim S-I, and Chang Y-J (2003) Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol Lett 25: 1869–1872.
Korn EL, Habermann JK, Upender MB, Ried T, and McShane LM (2004) Objective method of comparing DNA microarray image analysis systems. BioTechniques 6(6): 960–967.
Larkin JE, Frank BC, Gaspard RM, Duka I, Gavras H, and Quackenbush J (2004) Cardiac transcriptional response to acute and chronic angiotensin II treatments. Physiol Genomics 18: 152–166.
Lee ML, Kuo FC, Whitmore GA, and Sklar J (2000) Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridizations. Proc Natl Acad Sci USA 97: 9834–9839.
Li X-Q and Zhang D (2003) Gene expression activity and pathway selection for sucrose metabolism in developing storage root of sweetpotato. Plant Cell Physiol 44(6): 630–636.
Pavlidis P, Li Q, and Noble WS (2003) The effect of replication on gene expression microarray experiments. Bioinformatics 19(13): 1620–1627.
Qin L, Rueda L, Ali A, and Ngom A (2005) Spot detection and image segmentation in DNA microarray data. Appl Bioinformatics 4(1): 1–11.
Ringli C, Baumberger N, Diet A, Frey B, and Keller B (2002) ACTIN2 is essential for bulge site selection and tip growth during root hair development of Arabidopsis. Plant Physiol 129: 1464–1472.
Rosa GJM, Steibel JP, and Tempelman RJ (2005) Reassessing design and analysis of two-colour microarray experiments using mixed effects models. Comp Funct Genomics 6: 123–131.
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, and Quackenbush J (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34(2): 374–378.
Schena M, Shalon D, Davis RW, and Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470.
Shewry PR (2003) Tuber storage proteins. Ann Bot 91: 755–769.
Smyth GK and Speed TP (2003) Normalization of cDNA microarray data. Methods 31: 265–273.
Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology 3(1): Article 3.
Smyth GK (2005) Limma: linear models for microarray data. in: Gentleman R, Carey V, Dudoit S, Irizarry R, and Huber W (eds), Bioinformatics and computational biology solutions using R and bioconductor, pp. 397–420, New York: Springer.
Smyth GK, Michaud J, and Scott H (2005) The use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21(9): 2067–2075.
Spruill SE, Lu J, Hardy S, and Weir B (2002) Assessing sources of variability in microarray gene expression data. BioTechniques 33(4): 916–923.
Ullmannová V and Haškovec C (2003) The use of housekeeping genes (HKG) as an internal control for the detection of gene expression by quantitative real-time RT-PCR. Folia Biol 49: 211–216.
Volkov RA, Panchuk I, and Schöffl F (2003) Heat-stress-dependency and developmental modulation of gene expression: the potential of house-keeping genes as internal standards in mRNA expression profiling using real-time RT-PCR. J Exp Bot 54: 2343–2349.
Wei C, Jiangning L, and Bumgarner RE (2004) Sample size for detecting differentially expressed genes in microarray experiments. BMC Genomics 5: 87.
Wilson LA and Lowe SB (1973) The anatomy of the root system in West Indian sweetpotato (Ipomoea batatas (L.) Lam.) cultivars. Ann Bot 37: 633–643.
Wong ML and Medrano JF (2005) Real-time PCR for mRNA quantitation. BioTechniques 39(1): 75–85.
Woolfe J (1992) Sweetpotato: an untapped food resource. Cambridge University Press, New York, NY.
Yang YH, Buckley MJ, Dudoit S, and Speed TP (2002a) Comparison of methods for image analysis on cDNA microarray data. J Comput Graph Stat 11(1): 108–136.
Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, Wang S, Sharov V, Saeed AI, White J, Li J, Lee NH, Yeatman TJ, and Quackenbush J (2002b) Within the fold: assessing differential expression measures and reproducibility in microarray assays. {jtGenome Biol} {vn3}({sn11}): research 0062.1–0062.12
Yang YH and Speed T (2002) Design issues for cDNA microarray experiments. Nat Rev 3: 579–588.
Yuen T, Wurmbach E, Pfeffer R, Ebersole BJ, and Sealfon SC (2002) Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res 30(10): e48.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
McGregor, C.E., He, L., Ali, R.M. et al. The effect of replicate number and image analysis method on sweetpotato [Ipomoea batatas (L.) Lam.] cDNA microarray results. Plant Mol Biol Rep 23, 367–381 (2005). https://doi.org/10.1007/BF02788885
Issue Date:
DOI: https://doi.org/10.1007/BF02788885