Abstract
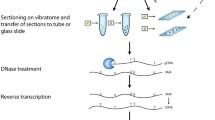
An essential facet of functional genomics is an efficient means of examining gene expression at single-cell resolution. We previously developed a high-throughput RT-PCR-based method for in situ localization of gene transcription products. As in all situ methods, it is assumed that the observed signal truly corresponds to the intended target, but this can be prone to false positives. Here we show that the labeled PCR product responsible for the actual histological signal may be recovered and directly analyzed using our method, thereby validating the histological result. We demonstrate that in situ reaction products for abundant (18S rRNA) and rare (Mt-ccs52, which encodes a cell cycle regulator) transcripts can be resolved by means of electrophoresis, blotting, and direct DNA sequencing. Using primers that span anMt-ccs52 intron and DNase treatment, we confirmed that the signal being detected was ultimately derived from mRNA. The ability to experimentally establish the specificity of an in situ signal after the fact enhances the high-throughput capability of the in-well RT-PCR method and further increases the utility of this technique as a powerful tool for functional genomics studies.
Similar content being viewed by others
Abbreviations
- DIG:
-
digoxigenin
- EST:
-
expressed sequence tags
- RT:
-
reverse transcription
References
Cebolla A, Vinardell JM, Kiss E, Olah B, Roudier F, Kondorosi A, and Kondorosi E (1999) The mitotic inhibitorccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plant. EMBO J 18: 4476–4484.
Koltai H and Bird DMcK (2000) High throughput cellular localization of specific plant mRNAs by liquid-phase in situ reverse transcription-polymerase chain reaction of tissue sections. Plant Physiol 123: 1203–1212.
Koltai H, Dhandaydham M, Thomas JF, Opperman C, and Brid DMcK (2001) Overlapping plant signal transduction pathways induced by a parasitic-nematode and a rhizobial endosymbiont. MPMI 14: 1168–1177.
Ewing BL, Hillier MC, Wendl MC, and Green P (1998) Base-calling of automated sequence traces using Phred. I. accuracy assessment. Genome Res 8: 175–185.
Schultze M and Kondorosi A (1998) Regulation of symbiotic root nodule development Annu Rev Genet 32: 33–57.
Somasegaran P and Hoben HJ (1994) Handbook for Rhizobia: methods in legumerhizobium technology. Springer-Verlag, New York.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Koltai, H., Bird, D.M. Recovery and sequence validation of the histological signal following in situ RT-PCR localization of plant gene transcripts. Plant Mol Biol Rep 20, 391–397 (2002). https://doi.org/10.1007/BF02772126
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF02772126