Summary
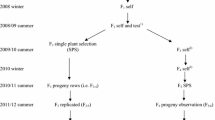
Somaclonal variation for quantitative traits could affect the practical utilization of regenerants in cotton improvement. Three groups of experimental lines were derived to analyze variation, including one control group from the explant-source cultivar and two groups of R3 somaclones from different R0’s (R0 = initial regenerant) free of observable chromosomal rearrangements. A three-environment field trial was conducted to evaluate group means, genetic variance, and line performance. Mean seedcotton yields of the somaclonal groups were reduced by 21 and 26% relative to the Coker 310 standard at two locations, but lint percentage and certain fiber properties were improved. Group-by-environment interactions were significant (P<0.05) for 10 of the 12 measured traits. Genetic variance tended to decrease in the somaclones, plant height being an exception. Line performance of the somaclones indicated that 50-boll weight, seed index, and fiber length did not reach the Coker 310 group means. These data suggest that genetic gain will be improved if regenerants of cotton are self-pollinated and the progenies evaluated for quantitative traits before crossing somaclones with the explantsource cultivar or other elite germplasm.
Similar content being viewed by others
References
Altman, D. W.; Busch, R. H. Random intermating before selection in spring wheat. Crop Sci. 24:598–601; 1984.
Altman, D. W.; Stelly, D. M.; Mitten, D. M., et al. Cytogenetic and quantitative variation in cotton somaclones. In: Abstracts VIIth International Congress on Plant Tissue and Cell Culture; 24–29 June 1990; Amsterdam. 1990:146.
Altman, D. W.; Stipanovic, R. D. Benedict, J. H. Terpenoid aldehydes in upland cottons. II. Genotype-environment interactions. Crop. Sci. 29:1451–1456; 1989.
Altman, D. W.; Stipanovic, R. D.; Mitten, D. M., et al. Interaction of cotton tissue culture cells andVerticillium dahliae. In Vitro Cell. Dev. Biol. 21:659–664; 1985.
Carver, B. F.; Johnson, B. B. Partitioning of variation derived from tissue culture of winter wheat. Theor. Appl. Genet. 78:405–410; 1989.
D’Amato, F.; Cytogenetics of plant cell and tissue cultures and their regenerants. CRC Critical Rev. Plant Sci. 3:73–112; 1985.
Davidonis, G. H.; Hamilton, R. H. Plant regeneration from callus tissue ofGossypium hirsutum L. Plant Sci. Lett. 32:89–93; 1983.
Demarly, Y. L’épigénique. Bull. Soc. Bot. Fr. Actual. Bot. 132:79–94; 1985.
Evans, D. A. Applications of somaclonal variation. Adv. Biotechnol. Proc. 9:203–223; 1988.
Finer, J. J.; McMullen, M. D. Transformation of cotton (Gossypium hirsutum L.) via particle bombardment. Plant Cell Rep. 8:586–589; 1990.
Gwyn, J. J.; Stelly, D. M. Method to evaluate pollen viability of upland cotton: tests with chromosome translocations. Crop Sci. 29:1165–1169; 1989.
Larkin, P. J.; Scowcroft, W. R. Somaclonal variation—a novel source of variability from cell cultures for plant improvement. Theor. Appl. Genet. 60:197–214; 1981.
Lee, M.; Phillips, R. L. The chromosomal basis of somaclonal variation. Ann. Rev. Plant Physiol. Plant Mol. Biol. 39:413–437; 1988.
Li, R.; Stelly, D. M.; Trolinder, N. L. Cytogenetic abnormalities in cotton (Gossypium hirsutum L.) cell cultures. Genome 32:1128–1134; 1989.
Phillips, R. L.; Kaeppler, S. M.; Peschke, V. M. Do we understand somaclonal variation? In: Nijkamp, H. J. J.; van der Plas, L. H. W.; van Aartrijk, J., eds. Progress in plant cellular and molecular biology: Dordrecht, The Netherlands:Kluwer; 1990:131–141.
Quisenberry, J. E.; Trolinder, N. L. Somaclonal variation in fiber properties of upland cotton. Agron. Abstr. p. 96; 1989.
Scowcroft, W. R. Somaclonal variation: the myth of clonal uniformity. In: Hohn, B.; Dennis, E. S., eds. Genetic flux in plants. Vienna:Springer-Verlag; 1985:217–245.
Sibi, M. Heredity of epigenetic-variant plants from culture in vitro. In: Lange, W.; Zeven, A. C.; Hogenboom, N. G., eds. Efficiency in plant breeding. Proceedings of the 10th Congress of European Association for Research on Plant Breeding. Wageningen, The Netherlands: Pudoc; 1984:196–198.
Stelly, D. M.; Altman, D. W.; Kohel, R. J., et al. Cytogenetic abnormalities of cotton somaclones from callus cultures. Genome 32:762–770; 1989.
Trolinder, N. L. Xhixian, C. Genotype specificity of the somatic embryogenesis response in cotton. Plant Cell Rep. 8:133–136; 1989.
Umbeck, P.; Johnson, G.; Barton, K., et al. Genetically transformed cotton (Gossypium hirsutum L.) plants. Bio/Technology, 5:263–266; 1987.
Umbeck, P.; Swain, W.; Yang, N.-S. Inheritance and expression of genes for kanamycin and chloramphenicol resistance in transgenic cotton plants. Crop Sci. 29:196–201; 1989.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Altman, D.W., Stelly, D.M. & Mitten, D.M. Quantitative trait variation in phenotypically normal regenerants of cotton. In Vitro Cell Dev Biol - Plant 27, 132–138 (1991). https://doi.org/10.1007/BF02632196
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02632196