Abstract
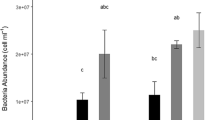
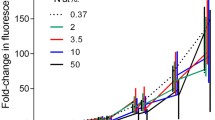
During routine [3H]thymidine incorporation measurements of environmental samples, significant amounts of radioactivity are often incorporated into macromolecules other than DNA. Although the percentage of nonspecific labeling varies both temporally and spatially, the cause(s) of these variations remain unknown. Correlations between the percent incorporated radioactivity in DNA and a variety of experimental and environmental parameters measured in the Alfia River, Crystal River, Medard Reservoir, and Bayboro Harbor were examined. The amount of radioactivity incorporated into DNA ranged from 6 to 95% (\(\bar x = 51 \pm 14\% \); n=121). Nonspecific labeling began immediately upon the addition of [3H]thymidine and was linear over time. Labeling patterns were independent of both the amount of thymidine added and cell-size fraction. A two year study of Bayboro Harbor indicated no conclusive relationship between nonspecific labeling and seasonality. The amount of radioactivity incorporated into DNA was inversely correlated with total rates of thymidine incorporation and a strong diurnal pattern was observed in the Crystal River. No consistent relationship was observed between labeling patterns and primary productivity, chlorophylla, particulate DNA, dissolved DNA, bacterial cell numbers, temperature, salinity, and dissolved organic carbon. The only relationship with dissolved inorganic nutrients (N and P) occurred in the Crystal River. In this phosphate limited river, the percent of radioactivity incorporated into DNA was positively correlated with phosphate concentrations. These results indicate that nonspecific labeling is not dependent on any one parameter but may be a function of many interacting environmental factors or a function of the specific ambient bacterial population.
Similar content being viewed by others
References
Armstrong FAJ, Sterns CR, Strickland JDH (1967) The measurement of upwelling and subsequent biological processes by means of the Technicon autoanalyzer and associated equipment, Deep Sea Res 14:381–389
Bendschneider K, Robinson RJ (1952) A new method for the determination of nitrite in seawater. Mar Res 11:87–96.
Brittain AM, Karl DM (1990) Catabolism of tritiated thymidine by aquatic microbial communities and incorporation of tritium into RNA and protein. Appl Environ Microbiol 56:1245–1254.
Carmen KR, Dobbs FC, Guckert JB (1988) Consequences of thymidine catabolism for estimates of bacterial production: An example from a coastal marine sediment. Limnol Oceanogr 33:1595–1606
Carmody JM, Herriott RM (1970) Thymine and thymidine uptake byHeamophilus influenza and the labeling of deoxyribonucleic acid. J Bacteriol 101:525–530
Carpenter EJ, Lively JS (1980) Review of estimates of algal growth using14C-tracer techniques. In: Falkowski PG (ed) Primary productivity in the sea. Brookhaven Symposium in Biology, No. 31. Plenum Press, New York, pp 161–178
David AW, Paul JH (1989) Enumeration and sizing of aquatic bacteria by use of a silicon-intensified target camera linked-image analysis system. J Microb Meth 9:257–266
DeFlaun MF, Paul JH, Jeffrey WH (1987) Distribution and molecular weight of dissolved DNA in subtropical estuarine and oceanic environments. Mar Ecol Prog Ser 38:65–73
Fallon RD, Newell SY (1986) Thymidine incorporation by the microbial community of standing deadSpartina alterniflora. Appl Environ Microbiol 52:1206–1208
Findley SEG, Meyer JL, Edwards RT (1984) Measuring bacterial production via rate of incorporation of3H-thymidine into DNA. J Microb Meth 2:57–72
Flannery MS (1989) Tampa and Sarasota Bays: Watersheds and tributaries: In: Estevez ED (ed) Tampa and Sarasota bays: Issues, resources, status, and management. NOAA estuary of the month seminar no. 7. US Dept Commerce
Fletcher WL, Ogle JK, Obery JL, Todd JM (1987) Water resources data-Florida water year 1986. Volume 3A. Southwest Florida surface water. USGS Water Data Report FL-86-3A, p 397
Fredericks AD, Sackett WM (1970) Organic carbon in the Gulf of Mexico. J Geophys Res 75: 2199–2206
Fuhrman JA, Azam F (1982) Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: Evaluation and field results. Mar Biol 66:109–120
Golterman HL, Clyme RS (1971) Methods for chemical analysis of fresh waters IBP Handbook No. 8, Blackwell Oxford, p 166
Grasshoff K, Johannsen J (1972) A new sensitive and direct method for the automatic determination of ammonia in seawater. J Cons Int Expl Mer 34:516–521
Hansen RB, Lowery HK (1983) Nucleic acid synthesis in oceanic microplankton from the Drake Passage, Antartica: Evaluation of steady state growth. Mar Biol 73:79–89
Hollibaugh JT (1988) Limitations of the [3H]thymidine method for estimating bacterial productivity due to thymidine metabolism. Mar Ecol Progr Ser 43:19–30
Holm-Hansen O, Riemann B (1978) Chlorophylla determination: Improvements in methodology. Oikos 30:438–448
Jeffrey WH, Paul JH (1986) Activity measurements of planktonic and microfouling communities in a eutrophic estuary. Appl Environ Microbiol 51:157–162
Jeffrey WH, Paul JH (1986) Activity of an attached and free-livingVibrio sp. as measured by thymidine incorporation, p-iodonitrotetrazolium reduction, and ATP/DNA ratios. Appl Environ Microbiol 51:150–156
Jeffrey WH, Paul JH (1988) Effect of 5-fluoro-2′-deoxyuridine on [3H]thymidine incorporation by bacterioplankton in the waters of southwest Florida, Appl Environ Microbiol 54:331–336
Jeffrey WH, Paul JH (1988) Underestimation of DNA synthesis by [3H]thymidine incorporation in marine bacteria, Appl Environ Microbiol 54:3165–3168
Karl DM (1982) Selected nucleic acid precursers in studies of aquatic microbial ecology. Appl Environ Microbiol 50:706–709
Kornberg A (1980) DNA replication WH Freeman, San Francisco
Lovell RC, Konopka A (1985) Primary and bacterial production in two dimictic Indian lakes. Appl Environ Microbiol 49:485–491
Moriarty DJW (1986) Measurement of bacterial growth rates in aquatic systems from rates of nucleic acid synthesis. Adv Microb Ecol 9:245–292
Moriarty DJW, Pollard PC (1982) Diel variation of bacterial productivity in seagrass (Zostera capricorni) beds measured by rate of thymidine incorporation into DNA. Mar Biol 72:165–173
Murphy J, Riley JP (1962) A modified single solution method for determination of phosphate in natural waters. Anal Chim Acta 27:31–36
O'Donovan GA (1990) Thymidine metabolism in bacteria. EOS 71:110
Paul JH, DeFlaun MF, Jeffrey WH, David AW (1988) Seasonal and diel variability in dissolved DNA and in microbial biomass and activity in a subtropical estuary. Appl Environ Microbiol 54:718–727
Paul JH, Jeffrey WH, David AW, DeFlaun MF, Cazares LH (1989) Turnover of extracellular DNA in eutrophic and oligotrophic freshwaters of Southwest Florida. Appl Environ Microbiol 55:1823–1828
Pollard PC (1987) Dialysis: A simple method for separating labeled bacterial DNA and tritiated thymidine from aquatic sediments. J Microbiol Meth 7:91–101
Riemann B (1984) Determining growth rates of natural assemblages of freshwater bacteria by means of3H-thymidine incorporation into DNA: Comments on methodology. Arch Hydrobiol Beith Ergebn Limnol 19:67–80
Riemann B, Sondergaard M (1984) Measurements of diet rates of bacterial secondary production in aquatic environments. Appl Environ Microbiol 47:632–638
Rivkin RB (1986) Incorporation of tritiated thymidine by eukaryotic microalgae. J Phycol 22:193–198
Rivkin RB, Voytek MA (1986) Cell division rates of eukaryotic algae measured by tritiated thymidine incorporation into DNA: Coincident measurements of photosynthesis and cell division of individual species of phytoplankton isolates from natural populations. J Phycol 22:199–205
Robarts RD, Wicks RJ (1989) [Methyl-3H]thymidine macromolecular incorporation and lipid labeling: Their significance to DNA labeling during measurements of aquatic bacterial growth rate. Limnol Oceanogr 34:213–222
Robarts RD, Wicks RJ, Sephton LM (1986) Spatial and temporal variations in bacterial macromolecular labelling with [methyl-3H]thymidine in a hypersaline lake. Appl Environ Microbiol 52:1368–1373
Servais PJ, Martinez J, Billen G, Vives-Rego J (1987) Determining [3H]thymidine incorporation into bacteriaplankton DNA: Improvement of the method by DNase treatment. Appl Environ Microbiol 53:1977–1979
Thorn PM, Ventullo RM (1988) Measurement of bacterial growth rates in subsurface sediments using incorporation of tritiated thymidine into DNA. Microb Ecol 16:3–16
Wetzel KP, Graf G (1984) On the use of different nucleic acid precursers for the measurement of microbial nucleic acid turnover. Arch Hydrobiol 19:59–65
Wicks RJ, Robarts RD (1987) The extraction and purification of DNA labled with [methyl-3H]thymidine in aquatic bacterial production sutdies. J Plankton Res 9:1159–1166
Zar JH (1984) Biostatistical analysis Prentice Hall Inc., Englewood Cliffs
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Jeffrey, W.H., Paul, J.H., Cazares, L.H. et al. Correlation of nonspecific macromolecular labeling with environmental parameters during [3H]Thymidine incorporation in the waters of southwest florida. Microb Ecol 20, 21–35 (1990). https://doi.org/10.1007/BF02543864
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF02543864