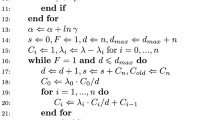Abstract
Markov chain Monte Carlo (MCMC) methods have become popular as a basis for drawing inference from complex statistical models. Two common difficulties with MCMC algorithms are slow mixing and long run-times, which are frequently closely related. Mixing over the entire state space can often be aided by careful tuning of the chain's transition kernel. In order to preserve the algorithm's stationary distribution, however, care must be taken when updating a chain's transition kernel based on that same chain's history. In this paper we introduce a technique that allows the transition kernel of the Gibbs sampler to be updated at user specified intervals, while preserving the chain's stationary distribution. This technique seems to be beneficial both in increasing efficiency of the resulting estimates (via Rao-Blackwellization) and in reducing the run-time. A reinterpretation of the modified Gibbs sampling scheme introduced in terms of auxiliary samples allows its extension to the more general Metropolis-Hastings framework. The strategies we develop are particularly helpful when calculation of the full conditional (for a Gibbs algorithm) or of the proposal distribution (for a Metropolis-Hastings algorithm) is computationally expensive.
Similar content being viewed by others
References
Abrams D I, Goldman A I, Launer C, Korvick J A, Neaton J D, Crane L R et al. (1994) Comparative trial of didanosine and zalcitabine in patients with human immunodeficiency virus infection who are intolerant or have failed zidovudine therapy. New England Journal of Medicine330: 657–662
Besag J, Green P J (1993) Spatial statistics and Bayesian computation (with discussion). J. Roy. Stat. Soc., Ser. B55: 25–37
Casella G, Robert C P (1996) Rao-Blackwellisation of sampling schemes. Biometrika83: 81–94
Damien P, Wakefield J, Walker S (1999) Gibbs sampling for Bayesian nonconjugate and hierarchical models using auxiliary variables. J. Roy. Stat. Soc., Ser. B61: 331–344
Gelfand A E, Sahu S K, Carlin B P (1995) Efficient parametrizations for normal linear mixed models. Biometrika82: 479–488
Gelfand A E, Smith A F M (1990) Sampling based approaches to calculating marginal densities. Journal of the American Statistical Association85: 398–409
Geyer C J, Thompson E A (1995) Annealing Markov chain Monte Carlo with applications to ancestral inference. J. Amer. Stat. Assoc.90: 909–920
Gilks W R, Roberts G O (1996) Strategies for improving MCMC. In: Gilks W R, Richardson S, Spiegelhalter D J (eds.) Markov Chain Monte Carlo in Practice, pp. 89–114. Chapman and Hall, London
Gilks W R, Roberts G O, Sahu S K (1998) Adaptive Markov chain Monte Carlo through regeneration. J. Amer. Stat. Assoc.93: 1045–1054
Hastings W K (1970) Monte Carlo sampling methods using Markov chains and their applications. Biometrika57: 97–109
Hills S E, Smith AFM (1992) Parametrization issues in Bayesian inference. In: Bernardo J M, Berger J O, Dawid, A P, Smith A F M (eds.) Bayesian statistics 4, pp. 641–649. Oxford University Press, Oxford
Hodges J S (1998) Some algebra and geometry for hierarchical models, applied to diagnostics (with discussion). J. Roy. Stat. Soc., Ser B60: 497–536
Kass R E, Carlin B P, Gelman A, Neal R (1998) Markov chain Monte Carlo in practice: a roundtable discussion. Amer. Stat.52: 93–100
Liu J S, Wong W H, Kong A (1995) Correlation structure and convergence rate of the Gibbs sampler with various scans. J. Roy. Statist. Soc. Ser. B57: 157–169
Mira A, Tierney L (2002) Efficiency and convergence properties of slice samplers. Scandinavian Journal of Stat.29: 1–12
Neal R M (1996) Sampling from multimodal distributions using tempered transitions. Stat. and Comp.6: 353–366
Sargent D J, Hodges J S (1997) Smoothed ANOVA with application to subgroup analysis. Research Report 97-002, Division of Biostatistics, University of Minnesota
Sargent D J, Hodges J S, Carlin B P (2000) Structured Markov chain Monte Carlo. Journal of the Computational and Graphical Statistics9: 217–234
Spiegelhalter D J, Thomas A, Best N, Gilks W R (1995) BUGS: Bayesian Inference Using Gibbs Sampling, Version 0.50. Technical report, Medical Research Council Biostatistics Unit, Institute of Public Health, Cambridge University
Swendsen R H, Wang J S (1987) Non-universal critical dynamics in Monte Carlo simulations. Phys. Rev. Letters58: 86–88
Tierney L, Mira A (1999) Some adaptive Monte Carlo methods for Bayesian inference. Statistics in Medicine18: 2507–2515
Author information
Authors and Affiliations
Additional information
Partial financial support from FAR 2002-3, University of Insubria is gratefully acknowledged.
Rights and permissions
About this article
Cite this article
Mira, A., Sargent, D.J. A new strategy for speeding Markov chain Monte Carlo algorithms. Statistical Methods & Applications 12, 49–60 (2003). https://doi.org/10.1007/BF02511583
Issue Date:
DOI: https://doi.org/10.1007/BF02511583