Abstract
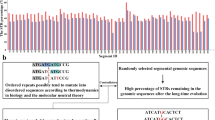
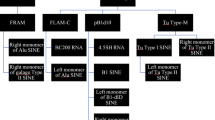
Most animal genomes contain a large number of short interspersed elements (SINEs) that have a composite structure and contain a region that is homologous to a tRNA. The majority of SINEs have been found to be derived from a tRNALys, being categorized as members of a superfamily of tRNALys-related SINEs. The consensus sequences of five SINEs that belong to this superfamily were aligned. It was found that, in the tRNA-unrelated region, there are two sequence motifs that are almost identical among these five SINEs and are at a distance of 10–11 nucleotides from each other. This observation suggests a common evolutionary origin of these SINEs and/or some function(s) for these motifs. Similar sequences were unexpectedly found to be present in the sequences complementary to the U5 regions of several mammalian retroviruses whose primer is a tRNALys. On the basis of these findings, we propose a possible model for the generation of SINEs whereby they are derived from a “strong stop DNA” with a primer tRNA that is an intermediate in the process of reverse transcription of certain retroviruses.
Similar content being viewed by others
References
Daniels GR, Deininger PL (1985) Repeat sequence families derived from mammalian tRNA genes. Nature (London) 317:819–822
Denison RA, Weiner AM (1982) Human U1 RNA pseudogenes may be generated by both DNA- and RNA-mediated mechanisms. Mol Cell Biol 2:815–828
Derse D, Dorn PL, Levy L, Stephens RM, Rice NR, Casey JW (1987) Characterization of equine infectious anemia virus long terminal repeat. J Virol 61:743–747
Endoh H, Okada N (1986) Total DNA transcription in vitro: A procedure to detect highly repetitive and transcribable sequences with tRNA-like structures. Proc Natl Acad Sci USA 83:251–255
Endoh H, Nagahashi S, Okada N (1990) A highly repetitive and transcribable sequence in tortoise genome is probably a retroposon. Eur J Biochem 189:25–31
Fasel N, Buetti E, Firzlaff J, Pearson K, Diggelmann H (1983) Nucleotide sequence of the 5′ noncoding region and part of thegag gene of mouse mammary tumor virus: identification of the 5′ splicing site for subgenomic mRNAs. Nucleic Acids Res 11:6943–6955
Gojobori T, Yokoyama S (1985) Rates of evolution of the retroviral oncogene of Moloney murine sarcoma virus and of its cellular homologues. Proc Natl Acad Sci USA 82:4198–4201
Kido Y, Aono M, Yamaki T, Matsumoto K, Murata S, Saneyoshi M, Okada N (1991) Shaping and reshaping of salmonid genomes by amplification of tRNA-derived retroposons during evolution. Proc Natl Acad Sci USA 88:2326–2330
Krayev AS, Markusheva TV, Kramerov DA, Ryskov AP, Skryabin KG, Bayev AA, Georgiev GP (1982) Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res 10:7461–7475
Lawrence CB, McDonnell DP, Ramsey WJ (1985) Analysis of repetitive sequence elements containing tRNA-like sequences. Nucleic Acids Res 13:4239–4252
Matsumoto K, Murakami K, Okada N (1986) Gene for lysine tRNA1 may be a progenitor of the highly repetitive and transcribable sequences present in the salmon genome. Proc Natl Acad Sci USA 83:3156–3160
Mochizuki K, Umeda M, Ohtsubo H, Ohtsubo E (1992) Characterization of a plant SINE, p-SINE1, in rice genomes. Jpn J Genet 67:155–166
Ohshima K, Koishi R, Matsuo M, Okada N (1993) Several SINEs in distant species may have originated from a common ancestral retrovirus: characterization of a squid SINE and a possible mechanism for generation of tRNA-derived retroposons. Proc Natl Acad Sci USA in press
Okada N et al (1991) Evolution of repetitive sequences. In: Evolution of Life. Osawa S, Honjo T (eds) Springer-Verlag, Tokyo, pp 175–186
Okada N (1991a) SINEs: Short interspersed repeated elements of the eukaryotic genome. Trends Ecol Evol 6:358–361
Okada N (1991b) SINEs. Curr Opin in Genet Dev, 1:498–504
Okada N (1990) Transfer RNA-like structure of the human Alu family: implication of its generation mechanism and possible functions. J Mol Evol 31:500–510
Power MD Marx PA, Bryant ML, Gardner MB, Barr PJ, Luciw PA (1986) Nucleotide sequence of SRV-1, a type D simian aquired immune deficiency syndrome retrovirus. Science 231:1567–1572
Rogers J (1985) The structure and evolution of retroposons. Int Rev Cytol 93:231–279
Saigo K (1986) Acopia primer pseudogen possibly generated by an abberant reverse transcription of acopia-related element inDrosophila. Nucleic Acids Res 14:7815
Sakamoto K, Okada N (1985) Rodent type 2 Alu family, rat identifier sequence, rabbit C family, and bovine or goat 73-bp repeat may have evolved from tRNA genes. J Mol Evol 22:134–140
Singer MF (1982) SINEs and LINEs: Highly repeated short and long interspersed sequences in mammalian genomes. Cell 28:433–434
Singer M, Berg P (1991) Genes and Genomes, University Science Books
Sonigo P, Barker C, Hunter E, Wain-Hobson S (1986) Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell 45:375–385
Spire B, Sire J, Zachar V, Rey F, Barré-Sinoussi F, Galibert F, Hampe A, Chermann J-C (1989) Nucleotide sequence of HIV1-NDK: a highly cytopathic strain of the human immunodeficiency virus. Gene 81:275–284
Ullu E, Tschudi C (1984) Alu sequences are processed 7SL RNA genes. Nature (London) 312:171–172
Weiner AM, Deininger PL, Efstratiadis A (1986) Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem 55:631–661
Weiner AM (1980) An abundant cytoplasmic 7S RNA is complementary to the dominant interspersed middle repetitive DNA sequence family in the human genome. Cell 22:209–218
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Okada, N., Ohshima, K. A model for the mechanism of initial generation of short interspersed elements (SINEs). J Mol Evol 37, 167–170 (1993). https://doi.org/10.1007/BF02407352
Issue Date:
DOI: https://doi.org/10.1007/BF02407352