Abstract
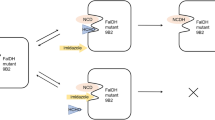
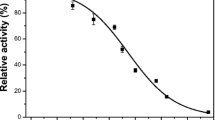
Molecular docking simulations were performed in this study to investigate the importance of both structural and catalytic zinc ions in the human alcohol dehydrogenase β2β2 on substrate binding. The structural zinc ion is not only important in maintaining the structural integrity of the enzyme, but also plays an important role in determining substrate binding. The replacement of the catalytic zinc ion or both catalytic and structural zinc ions with Cu2+ results in better substrate binding affinity than with the wild-type enzyme. The width of the bottleneck formed by L116 and V294 in the substrate binding pocket plays an important role for substrate entrance. In addition, unfavorable contacts between the substrate and T48 and F93 prevent the substrate from moving too close to the metal ion. The optimal binding position occurs between 1.9 and 2.4 Å from the catalytic metal ion.
Similar content being viewed by others
References
Barlow S, Rohl AL, O'Hare D. Molecular Mechanics Study of Oligomeric Models for Poly(ferrocenylsilanes) using the ESFF Forcefield. Chem Comm 257–260;1996.
Barlow S, Rohl, AL, Shi S, Freeman CM, O'hare D. Molecular Mechanics Study of Oligomeric Models for Poly(Ferrocenylsilanes) using the Extensible Systematic Forcefield (ESFF). J Am Chem Soc 118:7578–7592;1996.
Bogin O, Peretz M, Burstein Y.Thermoanaerobacter brockii alcohol dehydrogenase: Characterization of the active site metal and its ligand amino acids. Protein Sci 6:450–458;1997.
Brown DC, Collins KD. Dihydroorotase fromEscherichia coli. Substitution of Co(II) for the active site Zn(II). J Biol Chem 266:1597–1604;1991.
Crosas B, Allali-Hassani A, Eva Martínez S, Martras S, Persson B, Jörnvall H, Parés X, Farrés H. Molecular basis for differential substrate specificity in class IV alcohol dehydrogenases. J Biol Chem 275:25180–25187;2000.
Duester G, Farrés J, Felder MR, Holmes R, Höög J-O, Parés X, Plapp BV, Yin S-J, Jörnvall H. Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem Pharm 58:389–395;1999.
Edenberg HJ, Bosron WF. Alcohol dehydrogenases. In: Guengerich FP. Comprehensive Toxicology, vol. 3. New York. Pergamon Press 119–131;1997.
Eklund H, Nordström B, Zeppezauer E, Söderlund G, Ohlsson I, Boiwe T, Söderberg B-O, Tapia O, Brändén C-I, Åkeson Å. Three-dimensional structure of horse liver alcohol dehydrogenase at 2.4 Å resolution. J Mol Biol 102:27–59;1976.
Eklund H, Plapp BV, Samama J-P, Brändén C-I. Binding of substrate in a ternary complex of horse liver alcohol dehydrogenase. J Biol Chem 257:14349–14358;1982.
Eklund H, Horjales E, Vallee BL, Jörnvall H. Computer-graphic interpretations of residue exchanges between the α, β, and γ subunits of human-liver alcohol dehydrogenase class I isoenzymes. Eur J Biochem 167:185–193;1987.
Foglio MH, Duester G. Molecular docking studies on interaction of diverse retinol structures with human alcohol dehydrogenases predict a broad role in retinoid ligand synthesis. Biochim Biophys Acta 1432:239–250;1999.
Holland DR, Hausrath AC, Juers D, Matthews BW. Structural analysis of zinc substitutions in the active site of thermolysin. Protein Sci 4:1955–1965;1995.
Holmquist B, Vallee BL. Metal-coordinating substrate analogs as inhibitors of metalloenzymes. Proc Natl Acad Sci USA 76:6216–6220;1979.
Hurley TD, Bosron WF, Stone CL, Amzel LM. Structures of three human beta alcohol dehydrogenase variants. Correlations with their functional differences. J Mol Biol 239:415–29;1994.
Jörnvall H, Höög J-O. Nomenclature of alcohol dehydrogenases. Alcohol Alcohol 30:153–161;1995.
Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637;1983.
Kägi JHR, Vallee BL. The role of zinc in alcohol dehydrogenase. J Biol Chem 235:3188–3192;1960
Marschall H-U, Opperman UCT, Svensson S, Nordling E, Persson B, Höög J-O, Jörnvall H. Human liver class I alcohol dehydrogenase γγ isozyme: The sole cytosolic 3β-hydroxysteroid dehydrogenase of iso-bile acids. Hepatology 31:990–996;2000.
Neale AD, Scopes RK, Kelly JM, Wettenhall RE. The two alcohol dehydrogenases ofZymomonas mobilis. Purification by differential dye ligand chromatography, molecular characterization and physiological roles. Eur J Biochem 154:119–124;1986.
Niederhut MS, Gibbsons BJ, Perez-Miller S, Hurley TD. Three-dimensional structures of the three human class I alcohol dehydrogenases. Protein Sci 10:697–706;2001.
Nowak W, Wojtczak A, Cody V. Computer modeling with the ESFF forcefield of human dihydrofolate reductase ternary complex with NADPH and piritrexim (PTX) inhibitor. 5th Electronic Computational Chem Conference paper 46;1998.
Scopes RK. An iron-activated alcohol dehydrogenase. FEBS Lett 156:303–306;1983.
Svenssonm S, Some M, Lundsjö A, Helander A, Cronholm T, Höög J-O. Activities of human alcohol dehydrogenases in the metabolic pathways of ethanol and serotonin. Eur J Biochem 262:324–329;1999.
Svensson S, Höög J-O, Schneider G, Sandalova T. Crystal structures of mouse class II alcohol dehydrogenase reveal determinants of substrate specificity and efficiency. J Mol Biol 302:441–453;2000.
Vallee BL, Auld DS. Active-site zinc ligands and activated H2O of zinc enzymes. Proc Natl Acad Sci USA 87:220–224;1990.
Vallee BL, Auld DS. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry 29:5647–5659;1990.
Vallee BL, Auld DS. Cocatalytic zinc motifs in enzyme catalysis. Proc Natl Acad Sci USA 90:2715–2718;1993.
Vallee BL, Auld DS. New perspective on zinc biochemistry: Cocatalytic sites in multi-zinc enzymes. Biochemistry 32:6493–6500;1993.
Williamson VM, Paquin CE. Homology ofSaccharomyces cerevisiae ADH4 to an iron-activated alcohol dehydrogenase fromZymomonas mobilis. Mol Gen Genet 209:374–381;1987.
Yoshida A, Rzhetsky A, Hsu LC, Chang C. Human aldehyde dehydrogenase gene family. Eur J Biochem 251:549–557;1998.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Liu, HL., Ho, Y. & Hsu, CM. The effect of metal ions on the binding of ethanol to human alcohol dehydrogenase β2β2 . J Biomed Sci 10, 302–312 (2003). https://doi.org/10.1007/BF02256449
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02256449