Summary
In this paper we first show that the primary structure of U1 snRNA is homologous to that of tandem repeated pre-tRNA. Two sets of polymerase III promoter sites (the a and b boxes) are clearly recognisable at the appropriate positions in U1, although neither is functional; these sites occur in a degenerate form and their transcription is initiated by polymerase II. Moreover, several of the conserved subsequences of tRNAs that are not associated with transcription initiation (and supposedly are conserved because of their role in translation) are conserved in U1 as well, one of them being the pattern Py-Py-anticodon-Pu-Pu (for both anticodons of tandem tRNA).
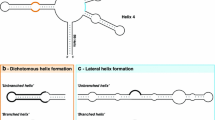
Second, we show that the secondary structure of U1 is apparently formed after fixation of the ‘B-hairpin loop’ by one of the associated proteins. If and only if this hairpin loop is fixed, a consensus secondary structure is produced by the minimisation-of-free-energy technique. Moreover, we show that this B-hairpin loop has been destabilised relatively recently in evolutionary time by deletions (e.g., in the polymerase III box). If we reinsert the deleted bases, the so constructed hypothetical “ancestral” molecule folds into the consensus secondary structure by unconstrained energy minimisation (i.e., without fixation of the B-loop).
Some features of the secondary structure of tandem repeated pre-tRNA are conserved in U1, but the overall structure has changed dramatically. Like tRNA, U1 has a cloverleaf-like structure, but its overall size has doubled. By comparing their secondary structures and by alignment of the sequences, we trace the local events associated with the global change in secondary structure (and apparently in the function of the molecule).
Finally, we discuss our results from the perspective of informatic prerequisites for heterarchical multilevel evolution.
Similar content being viewed by others
References
Altmann S (1975) Biosynthesis of transfer RNA inEscherichia coli. Cell 4:21–29
Auron PE, Rindone WP, Vary CPH, Celentano JJ, Vournakis JN (1982) Computer aided prediction of RNA secondary structure. Nucleic Acids Res 10:403–419
Bhat RA, Metz B, Thimmappaya B (1983) Organisation of noncontiguous coding components of adenovirus VAI RNA gene is strikingly similar to that of eukaryotic tRNA genes. Mol Cell Biol 3:1996–2005
Bralant C, Krol A, Ebel JP (1981) The conformation of chicken, rat and human U1a RNA's in solution. Nucleic Acids Res 9:841–858
Ciliberto G, Rangel G, Costanzo F, Dente L, Cortese R (1983) Common and interchangeable elements in the promoters of genes transcribed by polymerase III. Cell 32:725–733
Dayhoff MO, Schwartz RM, Chen HR, Hunt LT, Barker, WC, Orcutt BC (1981) Nucleic acid sequence database, vol 1. National Biomedical Research Foundation, Washington DC
Doolittle WF (1978) Genes in pieces: Were they ever together? Nature 272:581–582
Eigen M, Schuster P (1977) The hypercycle I. Naturwissenschaften 64:541–565
Eigen M, Schuster P (1978) The hypercycle II. Naturwissenschaften 65:7–41
Eigen M, Schuster P (1979) The hypercycle III. Naturwissenschaften 65:341–369
Eigen M, Winkler-Oswatitsch R (1981) Transfer-RNA: the early adaptor. Naturwissenschaften 68:217–292
Fuhrman SA, Engelke DR, Geidushek EP (1984) HeLa cell RNA polymerase III transcription factors. Biol Chem 259:1934–1943
Galli G, Hofstetter, H, Birnstiel M (1981) Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature 294:626–631
Garett RA, Douthwaite S, Noller HF (1981) Structure and role of 5S RNA-protein complexes in protein biosyntheses. TIBS 6:137–139
Hofstetter H, Kressman A, Birnstiel M (1981) A split promoter for eukaryotic tRNA gene. Cell 25:573–584
Hogeweg P, Hesper B (1984a) Energy directed folding of RNA sequences. Nucleic Acids Res 12:67–74
Hogeweg P, Hesper B (1984b) The alignment of sets of sequences and the construction of phyletic trees: an integrated method. J Mol Evol 20:175–186
Lund E, Dahlberg JE (1984) True genes for human U1 small nuclear RNA. Biol Chem 259:2013–2021
Marzluff WF, Brown DT, Lobo S, Wang SS (1983) Isolation and characterisation of two linked mouse U1-b small nuclear RNA genes. Nucleic Acids Res 11:6255–6270
Mount NSM, Steitz JA (1981) Sequence of U1 RNA fromDrosophila melanogaster: implications for U1 secondary structure and possible involvement in splicing. Nucleic Acids Res 9:6351–6368
Murphy MH, Baralle FE (1983) Directed, semisynthetic point mutational analysis of an RNA polymerase III. Nucleic Acids Res 11:7695–7700
Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in amino acid sequences of two proteins. J Mol Biol 48:443–453
Nussinov R, Jacobson AB (1980) Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc Natl Acad Sci USA 77:6309–6313
Nussinov R, Tinoco I (1981) Sequential folding of a messenger RNA molecule. J Mol Biol 151:519–533
Nussinov R, Pieczenik G, Griggs JR, Kleitman DJ (1978) Algorithms for loop matchings. SIAM J Appl Math 35:68–82
Nussinov R, Tinoco I, Jacobson AB (1982) Small changes in free energy assignments for unpaired bases do not affect predicted secondary structures in single stranded RNA. Nucleic Acids Res 10:341–349
Pipas JM, McMahon JE (1975) Method for predicting RNA secondary structure. Proc Natl Acad Sci USA 72:2017–2021
Queen CL, Korn LJ (1980) Computer analysis of nucleic acids and proteins. Methods Enzymol 65:595–609
Rohan RM, Ketner G (1983) Point mutations in the regulatory region of the human adenovirus Va1 gene. Biol Chem 258:11576–11581
Salser W (1977) Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harbor Symp Quant Biol 42:985–1102
Sellars PH (1974) On the theory of the computation of evolutionary distances. SIAM J Appl Math 26:787–793
Smith TF, Waterman MS, Fitch WM (1981) Comparative biosequence metrics. J Mol Evol 18:38–46
Studnicka GM, Rahn GM, Cummings IW, Salser WA (1978) Computer method for predicting the secondary structure of single stranded RNA. Nucleic Acids Res 5:3365–3387
Waterman MS, Smith TF, Beyer WA (1976) Some biological sequence metrics. Adv Math 20:267–287
Woese CR, Luersen CDP, Fox GE (1976) Sequence characterisation of 5S ribosomal RNA from eight Gram-positive prokaryotes. J Mol Evol 8:143–153
Zucker M, Striegler P (1981) Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res 9:133–148
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Hogeweg, P., Konings, D.A.M. U1 snRNA: The evolution of its primary and secondary structure. J Mol Evol 21, 323–333 (1985). https://doi.org/10.1007/BF02115650
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF02115650