Summary
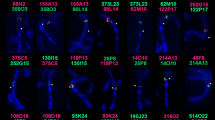
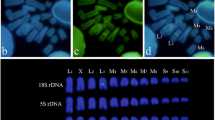
The Cervidae show one of the largest variations in chromosome number found within a mammalian family. The five species of the deer family which are the subject of this study vary in chromosome number from 2n=70 to 2n=6. Digestion with the restriction enzymes EcoRI, HpaII, HaeIII and MspI reveals that there is a series of highly repetitive sequences forming similar band patterns in the different species. To obtain information on the degree of homology among these conserved sequences we isolated a HpaII restriction fragment of approximately 990 base pairs from reindeer DNA. This DNA sequence was32P-labelled and hybridized by the Southern blot technique to DNAs cleaved with HpaII and HaeIII from the reindeer and four other Cervidae species. Hybridization to specific restriction fragments was recorded in all species. The patterns of hybridization showed a higher degree of similarity between reindeer, elk and roe deer than between reindeer and the Asiatic species (fallow deer and muntjac). Homologies are still present between the highly repetitive sequences of the five species despite the drastic reorganization that led to a change in chromosome number from 6 to 70.
Similar content being viewed by others
References
Baccus R, Ryman N, Smith MH, Reuterwall C, Cameron D (1983) Genetic variability and differentiation of large grazing mammals. J Mammal 64:109–120
Bedbrook JR, Jones J, O'Dell M, Thompson RD, Flavell RB (1980) A molecular description of telomeric heterochromatin inSecale species. Cell 19:545–560
Brykov VA, Volfson VG, Vorob'ev VI (1979) Genome structure and divergence of nucleotide sequences in echinodermata. Chromosoma 74:105–124
Flavell RB, Rimpau J, Smith DB (1977) Repeated sequence DNA relationships in four cereal genomes. Chromosoma 63:205–222
Gall JG, Atherton DD (1974) Satellite DNA sequences inDrosophila virilis. J Mol Biol 85:633–664
Jeffreys AJ, Flavell RA (1977) A physical map of the DNA regions flanking the rabbit β-globin gene. Cell 12:429–439
Lima-de-Faria A (1973) Equations defining the position of ribosomal cistrons in the eukaryotic chromosome. Nature (New Biol) 241:136–139
Lima-de-Faria A (1980) How to produce a human with 3 chromosomes and 1000 primary genes. Hereditas 93:47–73
Lima-de-Faria A, Pero R, Avanzi S, Durante M, Ståhle U, D'Amato F, Granström H (1975) Relation between ribosomal RNA genes and the DNA satellites ofPhaseolus coccineus. Hereditas 79:5–20
Lima-de-Faria A, Isaksson M, Olsson E (1980) Action of restriction endonucleases on the DNA and chromosomes ofMuntiacus muntjak. Hereditas 92:267–273
Maniatis T, Jeffrey A, Kleid DG (1975) Nucleotide sequence of the rightward operator of phage λ. Proc Natl Acad Sci USA 72:1184–1188
Musich PR, Brown FL, Maio JJ (1978) Mammalian repetitive DNA and the subunit structure of chromatin. Cold Spring Harbor Symp Quant Biol 42:1147–1160
Peacock WJ, Appels R, Dunsmuir P, Lohe AR, Gerlach WL (1977) Highly repeated DNA sequences: chromosomal localization and evolutionary conservatism. In: Brinkley BR, Porter KR (eds) Int Cell Biol 1976–1977. Rockefeller University Press, New York, pp 494–506
Peacock WJ, Lohe AR, Gerlach WL, Dunsmuir P, Dennis ES, Appels R (1978) Fine structure and evolution of DNA in heterochromatin. Cold Spring Harbor Symp Quant Biol 42:1121–1135
Pech M, Streeck RE, Zachau HG (1979) Patchwork structure of a bovine satellite DNA. Cell 18:883–893
Rigby PWJ, Dieckmann M, Rhodes C, Berg P (1977) Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol 113:237–251
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Stephenson EC, Erba HP, Gall JG (1981) Histone gene clusters of the newtNotophthalmus are separated by long tracts of satellite DNA. Cell 24:639–647
Waalwijk C, Flavell RA (1978) MspI, an isoschizomer of HpaII which cleaves both unmethylated and methylated HpaII sites. Nucleic Acids Res 5:3231–3236
Yasmineh WG and Yunis JJ (1970) Localization of mouse satellite DNA in constitutive heterochromatin. Exp Cell Res 59:69–75
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Lima-de-Faria, A., Arnason, U., Widegren, B. et al. Conservation of repetitive DNA sequences in deer species studied by Southern blot transfer. J Mol Evol 20, 17–24 (1984). https://doi.org/10.1007/BF02101981
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF02101981