Summary
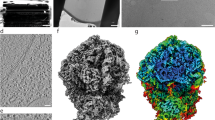
Multivariate statistical analysis and classification techniques are powerful tools in sorting noisy electron micrographs of single particles according to their principal features, enabling one to form average images with an enhanced signal-to-noise ratio and a better reproducible resolution. We apply this methodology here to determining the characteristic views of the large (50S) ribosomal subunits from the eubacteriumEscherichia coli and the archaebacteriaMethanococcus vannielii, Sulfolobus solfataricus, andHalobacterium marismortui. Average images were obtained of the subunit in the common crown and kidney projections, but views of the particle in orientations intermediate between these two extremes were also elucidated for all species. These averages show reproducible detail of up to 2.0 nm resolution, thus enabling the visualization and interspecies comparison of many structural features as a first step toward comparing the actual three-dimensional structures. Our results disprove evolutionary lineages recently postulated on the basis of electron microscopical images of ribosomal subunits.
Similar content being viewed by others
References
Bijlholt MMC, van Heel MG, van Bruggen EFJ (1982) Comparison of 4×6-meric hemocyanins from three different arthropods using computer alignment and correspondence analysis. J Mol Biol 161:139–153
Boekema EJ, Berden JA, van Heel M (1986) Structure of mitochondrial F1-ATPase studied by electron microscopy and image processing. Biochim Biophys Acta 851:353–360
Brock TD, Brock KM, Belley RT, Weiss RL (1972)Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Microbiol 84:54–68
Cowgill CA, Nichols BG, Kenny JW, Butler P, Bradbury EM, Traut RR (1984) Mobile domains in ribosomes revealed by proton nuclear magnetic resonance. J Biol Chem 259:15257–15263
Elhardt D, Boeck, A (1982) An in vitro polypeptide synthesizing system from methanogenic bacteria: sensitivity to antibiotics. Mol Gen Genet 188:128–134
Frank J, van Heel M (1982) Correspondence analysis of aligned images of biological particles. J Mol Biol 161:134–137
Frank J, Verschoor A, Boublik M (1982) Multivariate statistical analysis of ribosome electron micrographs. J Mol Biol 161: 107–133
Harauz G, Boekema E, van Heel M (1987) Statistical image analysis of electron micrographs of ribosomal subunits. Methods Enzymol (in press)
Henderson E, Oakes M, Clark MW, Lake JA, Matheson AT, Zillig W (1984) A new ribosome structure. Science 225:510–512
Hindennach I, Stoeffler G, Wittmann HG (1971) Isolation of proteins from 30S ribosomal subunits ofEscherichia coli. Eur J Biochem 23:7–11
Kiselev NA, Stel'mashchuk VYa, Orlova EV, Vasiliev VD, Selivanova OM (1982) Strand-like structures and their three-dimensional organization in the large subunit of theEscherichia coli ribosome. Mol Biol Rep 8:191–197
Kiselev NA, Orlova EV, Stel'maschuk VYa, Vasiliev VD, Selivanova OM, Kosykh VP, Pustovskikh AI, Kirichuk VS (1983) Computer averaging of 50S ribosomal subunit electron micrographs. J Mol Biol 169:345–350
Korn AP, Spitnik-Elson P, Elson D, Ottensmeyer FP (1983) Specific visualization of ribosomal RNA in the intact ribosome by electron spectroscopic imaging. Eur J Cell Biol 31: 334–340
Lake JA (1976) Ribosome structure determined by electron microscopy ofEscherichia coli small subunits, large subunits and monomeric ribosomes. J Mol Biol 105:131–159
Lake JA, Henderson E, Clark MW, Matheson AT (1982) Mapping evolution with ribosome structure: intralinege constancy and interlineage variation. Proc Natl Acad Sci USA 79: 5948–5952
Lake JA, Henderson E, Oakes M, Clark MW (1984) Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci USA 81:3786–3790
Lake JA, Clark MW, Henderson E, Fay SP, Oakes M, Scheinman A, Thornber JP, Mah RA (1985) Eubacteria, halobacteria, and the origin of photosynthesis: the photocytes. Proc Natl Acad Sci USA 82:3716–3720
Lebart L, Morineau A, Tabard N (1977) Techniques de la description statistique. Dunod, Paris
Lebart L, Morineau A, Warwick KM (1984) Multivariate descriptive statistical analysis. John Wiley & Sons, New York
Liljas A, Thirup S, Matheson AT (1986) Evolutionary aspects of ribosome-factor interactions. Chem. Scr 26B:109–119
Luehrmann R (1975) Identifizierung der Proteine S1, S13, und S18 als Komponenten der 30S ribosomalen Codon-Bindestelle. Dissertation. Universitaet Muenster, FRG
Matheson AT (1985) Ribosomes of archaebacteria. In: Woese CR, Wolfe RS (eds) The bacteria, vol 8. Academic Press, New York, pp 345–377
Meisenberger O, Pilz I, Stoeffler-Meilicke M, Stoeffler G (1984) Small angle X-ray study of the 50S ribosomal subunit ofEscherichia coli: a comparison of different models. Biochim Biophys Acta 781:225–233
Mevarech M, Eisenberg H, Neumann E (1977) Malate dehydrogenase isolated from extremely halophilic bacteria of the dead sea. 1. Purification and molecular characterization. Biochemistry 16:3781–3785
Noll M, Hapke B, Schreier MH, Noll H (1973) Structural dynamics of bacterial ribosomes. I. Characterization of vacant couples and their relation to complexed ribosomes. J Mol Biol 75:281–294
Oettl H, Hegerl R, Hoppe W (1983) Three-dimensional reconstruction and averaging of 50S ribosomal subunits ofEscherichia coli from electron micrographs. J Mol Biol 163:431–450
Olsen GJ, Pace NR, Nuell M, Kaine BP, Gupta R, Woese CR (1985) Sequence of the 16S RNA gene from the thermoacidophilic archaebacteriumSulfolobus solfataricus and its evolutionary implications. J Mol Evol 22:301–307
Schmid G, Boeck A (1982a) The ribosomal protein composition of the archaebacteriumSulfolobus. Mol Gen Genet 185: 498–501
Schmid G, Boeck A (1982b) The ribosomal protein composition of five methanogenic bacteria. Zentralbl Bakteriol Hyg, I Abt Orig C 3:347–353
Shevack A, Gewitz HS, Hennemann B, Yonath A, Wittmann HG (1985) Characterization and crystallization of ribosomal particles fromHalobacterium marismortui. FEBS Lett 184:68–71
Spiess E (1979) Structure of the 50S ribosomal, subunit fromEscherichia coli. Investigation of the intact subunit and core particles by electron microscopy and analogue image processing. Eur J Cell Biol 19:120–130
Stoeffler G, Stoeffler-Meilicke M (1984) Immunoelectron microscopy of ribosomes. Annu Rev Biophys Bioeng 13:303–330
Stoeffler G, Stoeffler-Meilicke M (1986) Electron microscopy of archaebacterial ribosomes. Syst Appl Microbiol 7:123–130
Stoeffler G, Lotti M, Noah M, Stoeffler-Meilicke M (1984) The localization of proteins L5 and L10 on the surface of 50S subunits ofEscherichia coli. In: Csanady A, Roehlich P, Szabo D (eds) Proceedings of the 8th European Congress on Electron Microscopy. Budapest, p 1551
Stoeffler-Meilicke M, Stoeffler G, Odom OW, Zinn A, Kramer G, Hardesty B (1981) Localization of 3′ ends of 5S and 23S rRNAs in reconstituted subunits ofEscherichia coli ribosomes. Proc Natl Acad Sci USA 78:5538–5542
Stoeffler-Meilicke M, Boehme C, Strobel O, Boeck A, Stoeffler G (1986) Structure of ribosomal subunits ofM. vannielii: ribosomal morphology as a phylogenetic marker. Science 231: 1306–1308
Tischendorf GW, Zeichardt H, Stoeffler G (1974) Determination of the location of proteins L14, L17, L18, L19, L22 and L23 on the surface of the 50S ribosomal subunit fromEscherichia coli by immune electron microscopy. Mol Gen Genet 134:187–208
van Heel M (1984a) Multivariate statistical analysis of images of biological macromolecules: powerful new methods. In: Csanády Á, Röhlich P, Szabó D (eds) Proceedings of the 8th European Congress on Electron Microscopy. Programme Committee, Budapest, p 1317
van Heel M (1984b) Multivariate statistical classification of noisy images (randomly oriented biological macromolecules). Ultramicroscopy 13:165–183
van Heel M (1987a) Similarity measures between images. Ultramicroscopy 21:95–100
van Heel M (1987b) Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy 21:111–124
van Heel M, Frank J (1981) Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy 6:187–194
van Heel M, Keegstra W (1981) IMAGIC: a fast, flexible and friendly image analysis software system. Ultramicroscopy 7: 113–130
van Heel M, Stoeffler-Meilicke M (1985) Characteristic views ofE. coli andB. stearothermophilus 30S ribosomal subunits in the electron microscope. EMBO J 4:2389–2395
van Heel M, Bretaudiere J-P, Frank J (1982) Classification and multi-reference alignment of images of macromolecules. In: Proceedings of the 10th International Congress on Electron Microscopy. Deutsche Gesellschaft fuer Elektronenmikroskopie, Frankfurt, p 563
Verschoor A, Frank J, Boublik M (1985) Investigation of the 50S ribosomal subunit by electron microscopy and image analysis. J Ultrastruct Res 92:180–189
Wittmann HG (1983) Architecture of prokaryotic ribosomes. Annu Rev Biochem 52:35–65
Woese CR, Olsen GJ (1986) Archaebacterial phylogeny: perspectives on the urkingdoms. Syst Appl Microbiol 7:161–177
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Harauz, G., Stoeffler-Meilicke, M. & van Heel, M. Characteristic views of prokaryotic 50S ribosomal subunits. J Mol Evol 26, 347–357 (1987). https://doi.org/10.1007/BF02101154
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02101154