Summary
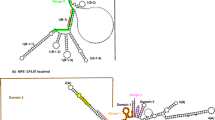
In order to determine why the aphid 28S rRNA lacks the hidden break otherwise found in insects, the structure of the region of the aphid ribosomal DNA (rDNA) corresponding to the gap region, which in other insect rDNA transcripts is excised posttranscriptionally, was studied. Sequence comparison suggested that, in contradistinction to what is found in rDNA transcripts of other insects, a stem-loop structure formed in this region of the aphid rDNA transcript is not AU-rich. Nor did the loop of the aphid molecule contain the UAAU tract that can be a signal for the introduction of the hidden break, suggesting that in this particular region the aphid 28S rRNA resembles 28S rRNAs of deuterostomes, which do not contain the hidden break.
Similar content being viewed by others
References
Benton WD, Davis RD (1977) Screening λgt recombinant clones by hybridization to single plaques in situ. Science 196:180–182
Buchner P (1965) Endosymbiosis of animals with plant micro-organisms. Interscience, New York
Clark CG, Tague BW, Ware VC, Gerbi SA (1984)Xenopus laevis 28S ribosomal RNA: a secondary structure model and its evolutionary and functional implications. Nucleic Acids Res 12:6197–6220
Delanversin G, Jacq B (1983) Molecular biology—sequence of the central break region ofDrosophila 26S precursor ribosomal RNA. C R Acad Sci Paris, Ser III 296:1041–1044
Erdmann VA, Wolters J (1986) Collection of published 5S, 5.8S and 4.5S ribosomal RNA sequences. Nucleic Acids Res 14: r1-r59
Fujiwara H, Ishikawa H (1982) Primary and secondary structures ofTetrahymena and aphid 5.8S rRNAs: structural features of 5.8S rRNA which interacts with the 28S rRNA containing the hidden break. Nucleic Acids Res 10:5173–5182
Fujiwara H, Ishikawa H (1986) Molecular mechanism of introduction of the hidden break into the 28S rRNA of insects: implication based on structural studies. Nucleic Acids Res 14:6393–6401
Gutell RR, Fox GE (1988) A compilation of large subunit RNA sequences presented in a structural format. Nucleic Acids Res 16:r175-r269
Hattori M, Sakaki Y (1986) Dideoxy sequencing method using denatured plasmid template. Anal Biochem 152:232–238
Henikoff S (1984) Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28:351–359
Ishikawa H (1977) Evolution of ribosomal RNA. Comp Biochem Physiol 58:1–7
Ishikawa H, Newburgh RW (1972) Studies of the thermal conversion of 28S RNA ofGalleria mellonella (L.) to an 18S product. J Mol Biol 64:135–144
Kossel H, Natt E, Strittmatter G, Fritzsche E, Gozdzicka-Jozefiak A, Przbyl D (1985) In: van Vioten-Doting L, Groot GSP, Hall TC (eds) Molecular form and function of the plant genome. Plenum, New York, pp 183–198
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning. Cold Spring Harbor Press, New York
Pavlakis GN, Jordan BR, Wurst RM, Vournakis JN (1979) Sequence and secondary structure ofDrosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res 7:2213–2238
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Ware VC, Tague BW, Clark CG, Gourse RL, Brand RC, Gerbi SA (1983) Sequence analysis of 28S ribosomal DNA from the amphibianXenopus laevis. Nucleic Acids Res 11:7795–7817
Ware VC, Renkawitz R, Gerbi SA (1985) rRNA processing: removal of only nineteen bases at the gap between 28Sα and 28Sβ rRNAs inSciara coprophila. Nucleic Acids Res 13: 3581–3597
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ogino, K., Eda-Fujiwara, H., Fujiwara, H. et al. What causes the aphid 28S rRNA to lack the hidden break?. J Mol Evol 30, 509–513 (1990). https://doi.org/10.1007/BF02101106
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02101106