Abstract
About one third of streptomycin resistance inEscherichia coli is mediated by APH-(3″). This enzyme is encoded by the plasmid pBP1 in 80 % of all streptomycin resistant strains tested. PBP1, which in addition mediates sulfonamide resistance, has been found to be disseminated inEscherichia coli strains all over the world. It has a molecular weight of 4.0 megadalton and does not seem to be disadvantageous for the metabolism of the bacterial cell. The reason for the slow decrease of resistance to streptomycin and sulfonamide in clinical isolates, despite the restricted use of these drugs, is presumably the survival of bacteria harbouring pBP1 which have been selected by streptomycin and sulfonamides in the early days of chemotherapy.
Similar content being viewed by others
References
Paul-Ehrlich-Gesellschaft für Chemotherapie e.V., Arbeitsgemeinschaft Resistenz: Resistenz klnischer Isolate einigerEnterobacteriaceae sowiePseudomonas aeruginosa, Staphylococcus aureur undStreptococcus faecalis gegenüber Chemotherapeutika. Ergebnisse einer überregionalen Studie aus den Jahren 1980 und 1981. Infection 1982, 10: 310–314.
Wiedemann, B., Weppelmann, G.: Amino-glycosidantibiotikamodifizierende Enzyme. Immunität und Infektion 1981, 9: 106–112.
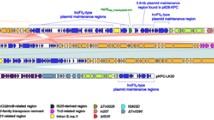
Van Treeck, U., Schmidt, F., Wiedemann, B.: Molecular nature of a streptomycin and sulfonamide resistance plasmid (pBP1) prevalent in clinicalEscherichia coli strains and integration of an ampicillin resistance transposon (TnA). Antimicrobial Agents and Chemotherapy 1981, 19: 371–380.
Kawabe, H., Tanaka, T., Mitsuhashi, S.: Streptomycin and spectinomycin resistance mediated by plasmids. Antimicrobial Agents and chemotherapy 1978, 13: 1031–1035.
Grinter, U., Barth, P. T.: Characterization of SmSu plasmids by restriction endonuclease cleavage and compatibility testing. Journal of Bacteriology 1976, 128: 394–400.
Grinsted, J., Saunders, J. R., Ingram, L. C., Sykes, R. B., Richmond, M. H.: Properties of an R factor which originated inPseudomonas aeruginosa 1822. Journal of Bacteriology 1972, 110: 529–537.
Achtman, M., Willetts, N., Clark, A. J.: Beginning a genetic analysis of conjugational transfer determined by the F factor inEscherichia coli by Isolation and characterization of transfer defective mutants. Journal of Bacteriology 1971, 106: 529–538.
Mise, K., Arber, W.: Plaque-forming transducing bacteriophage P1 derivatives and their behaviour in lysogenic conditions. Virology 1976, 69: 191–205.
Kushner, S. R.: An improved method for transformation ofEscherichia coli with ColE1 derived plasmids. In: Boyer, H. W., Nicosia, S. (ed.): Proceedings of the International Symposium on Genetic Engeneering. Elsevier Biomedical Press, Amsterdam, 1978, p. 17–23.
Guerry, P., van Embden, J. D. A., Falkow, S.: Molecular nature of two nonconjugative plasmids carrying drug resistance genes. Journal of Bacteriology 1974, 117: 619–630.
Bolivar, F., Rodriguez, R. L., Greene, P. J., Betlach, M. C., Heyneker, H. L., Boyer, H. W., Corsa, J., Falkow, S.: Construction and characterization of new cloning vehicles. II. A multi-purpose cloning system. Gene 1977, 2: 95–113.
Hansen, J. B., Olsen, R. H.: Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. Journal of Bacteriology 1978, 135: 227–238.
Casse, F., Boucher, C., Julliot, J. S., Michel, M., Dénarié, J.: Identification and characterization of large plasmids inRhizobium meliloti using agarose gel electrophoresis. Journal of General Microbiology 1979, 113: 229–242.
Kupersztoch-Portnoy, Y. M., Lovett, M. A., Helinski, D. R.: Strand and site specifity of the relaxation event for the relaxation complex of the antibiotic resistance plasmid R6K. Biochemistry 1974, 13: 5484–5490.
Meyers, J. A., Sanchez, D., Eiwell, L. P., Falkow, S.: Simple agarose electrophoresis method for the identification and characterization of plasmid deoxyribonucleic acid. Journal of Bacteriology 1976, 127: 1529–1537.
Sutcliffe, J. G.: pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Research 1978, 5: 2721–2728.
Elkhouly, A. E., Wiedemann, B.: Nature of streptomycin resistance in enterobacteria from faeces of streptomycin-treated patients in Egypt. Chemotherapy 1980, 26: 268–275.
Rubens, L., Heffron, F., Falkow, S.: Transposition of a plasmid deoxyribonucleic acid sequence that mediates ampicillin resistance: independence from hostrec functions and orientation of insertion. Journal of Bacteriology 1976, 128: 425–434.
Swedberg, G., Sköld, O.: Plasmid-borne sulfonamide resistance determinants studied by restriction enzyme analysis. Journal of Bacteriology 1983, 153: 1228–1237.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Korfmann, G., Lüdtke, W., van Treeck, U. et al. Dissemination of streptomycin and sulfonamide resistance by plasmid pBP1 inEscherichia coli . Eur. J, Clin. Microbiol. 2, 463–468 (1983). https://doi.org/10.1007/BF02013905
Issue Date:
DOI: https://doi.org/10.1007/BF02013905