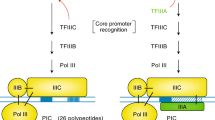
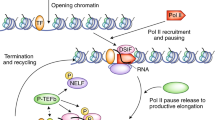
Summary
The basic components required for eukaryotic gene transcription have been highly conserved in evolution. Structural and functional homology has now been documented among promoters, promoter factors, regulatory proteins, and RNA polymerases from eukaryotes as diverse as yeast and mammals. The ability of these proteins and DNA sequences to function across phylogenetic boundaries demonstrates that common molecular mechanisms underlie gene control in all eukaryotic cells, and provides the basis for powerful new approaches to the study of eukaryotic gene transcription.
Similar content being viewed by others
Literature
Allison, L. A., Wong, J. K. C., Fitzpatrick, V. D., Moyle, M., and Ingles, C. J., The C-terminal domain of the largest subunit of RNA polymerase II ofSaccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Molec. cell. Biol.8 (1988) 321–329.
Allison, L. A., Moyle, M., Shales, M., and Ingles, C. J., Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell42 (1985) 599–610.
Beato, M., Gene regulation by steroid hormones. Cell56 (1989) 335–344.
Beggs, J. D., van den Berg, J., van Ooyen, A., and Weissmann, C., Abnormal express of chromosomal rabbit β-globin gene inSaccharomyces cerevisiae. Nature283 (1980) 835–840.
Benoist, C., O'Hara, K., Breathnach, R., and Chambon, P., The ovalbumin gene: sequence and putative control regions. Nucl. Acids Res.8 (1980) 127–142.
Berg, J., Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. Proc. natl Acad. Sci. USA85 (1988) 99–102.
Bishop, J. M., The molecular genetics of cancer. Science235 (1987) 305–311.
Bohmann, D., Bos, T. J., Admon, A., Nishimura, T., Vogt, P. K., and Tjian, R., Human protooncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science238 (1987) 1386–1392.
Brandl, C. J., and Struhl, K., Yeast GCN4 transcriptional activator protein interacts with RNA polymerase II in vitro. Proc. natl Acad. Sci. USA86 (1989) 2652–2656.
Buratowski, S., Hahn, S., Guarente, L., and Sharp, P. A., Five intermediate complexes in transcription initiation by RNA polymerase II. Cell56 (1989) 549–561.
Buratowski, S., Hahn, S., Sharp, P. A., and Guarente, L., Function of a yeast TATA element binding protein in a mammalian transcription system. Nature334 (1988) 37–42.
Burshell, A., Stathis, P. A., Do, Y., Miller, S. C., and Feldman, D., Characterization of an estrogen-binding protein in the yeastSaccharomyces cerevisiae. J. biol. Chem.259 (1984) 3450–3456.
Cavallini, B., Huet, J., Plassat, J.-L., Sentenac, A., Egly, J.-M., and Chambon, P., A yeast activity can substitute for the HeLa cell TATA box factor. Nature334 (1988) 77–80.
Chandler, V. L., Maler, B. A., and Yamamoto, K. R., DNA sequences bound specifically by the glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell33 (1983) 489–499.
Chodosh, L. A., Olesen, J., Hahn, S., Baldwin, A. S., Guarente, L., and Sharp, P. A., A yeast and a human CCAAT-binding protein have heterologous subunits that are functionally interchangeable. Cell53 (1988) 25–35.
Chodosh, L. A., Baldwin, A. S., Carthew, R. C., and Sharp, P. A., Human CCAAT-binding proteins have heterologous subunits. Cell53 (1988) 11–24.
Corden, J. L., Cadena, D. L., Ahearn, J. M., Jr., and Dahmus, M. E., A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc. natl Acad. Sci. USA82 (1985) 7934–7938.
Courey, A. J., and Tjian, R., Analysis of Sp1 in vivo reveals multiple transcription activation domains, including a novel glutamine-rich activation motif. Cell55 (1988) 887–898.
Evans, R. M., The steroid and thyroid hormone receptor superfamily. Science240 (1988) 889–895.
Fire, A., Samuels, M., and Sharp, P., Interactions between RNA polymerase II, factors, and template leading to accurate transcription. J. biol. Chem.259 (1984) 2509–2516.
Fischer, J. A., Giniger, E., Maniatis, T., and Ptashne, M., GAL4 activates transcription inDrosophila. Nature332 (1988) 853–856.
Freedman, L. P., Luisi, B. F., Korzun, Z. R., Basavappi, R., Sigler, P. K. B., and Yamamoto, K. R., The function and structure of the metal coordination sites within the glucocorticoid receptor DNA binding domain. Nature334 (1988) 543–546.
Gill, G., and Ptashne, M., Mutants of GAL4 protein altered in an activation function. Cell51 (1987) 121–126.
Gill, G., and Ptashne, M., Negative effect of the transcriptional activator GAL4. Nature334 (1988) 721–724.
Giniger, E., and Ptashne, M., Transcription in yeast activated by a putative amphiphathic α-helix linked to a DNA binding unit. Nature330 (1987) 670–672.
Godowski, P. J., Picard, D., and Yamamoto, K. R., Signal transduction and transcriptional regulation by glucocorticoid receptor-lexA fusion proteins. Science241 (1988) 812–816.
Green, S., and Chambon, P., Nuclear receptors enhance our understanding of transcription regulation. Trends Genet.4 (1988) 309–314.
Greenleaf, A. L., Borsett, L. M., Jiamachello, P. F., and Coulter, D. E., α-Amanitin resistantD. melanogaster with an altered RNA polymerase II. Cell18 (1979) 613–622.
Greenleaf, A. L., Amanitin-resistant RNA polymerase II mutations are in the enzyme's largest subunit. J. biol. Chem. (1983) 13403–13406.
Grosschedl, R., and Birnstiel, M. L., Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc. natl Acad. Sci. USA77 (1980) 7102–7106.
Guarente, L., UASs and enhancers: common mechanism of transcription activation in yeast and mammals. Cell52 (1988) 303–305.
Guarente, L., Regulatory proteins in yeast. A. Rev. Genet.21 (1987) 425–452.
Guarente, L., and Hoar, E., Upstream activation sites of the CYC1 gene ofSaccharomyces cerevisiae are active when inverted but not when placed downstream of the ‘TATA box’. Proc. natl Acad. Sci. USA81 (1984) 7860–7864.
Guarente, L., Lalonde, B., Gifford, B., and Alani, E., Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene ofS. cerevisiae. Cell36 (1984) 503–511.
Guarente, L., Yocum, R., and Gifford, P., A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc. natl Acad. Sci. USA79 (1982) 7410–7414.
Hahn, S., and Guarente, L., Yeast HAP2 and HAP3: Transcriptional activators in a heteromeric complex. Science240 (1988) 317–321.
Harshman, K. D., Moye-Rowley, W.S., and Parker, C. S., Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell53 (1988) 321–330.
Hawley, D. K., and Roeder, R. G., Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J. biol. Chem.260 (1985) 8163–8172.
Hochschild, A., Irwin, N., and Ptashne, M., Repressor structure and the mechanism of positive control. Cell32 (1983) 319–325.
Hollenberg, S. M., and Evans, R. M., Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell55 (1988) 899–906.
Hope, J. A., and Struhl, K., Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell46 (1986) 885–894.
Horikoshi, M., Carey, M. F., Kakidani, H., and Roeder, R. G., Mechanisms of action of a yeast activator: direct effect of GAL4 derivatives on mammalian TFIID-promoter interactions. Cell54 (1988) 665–669.
Jacob, F., and Monod, J., Genetic regulatory mechanisms in the synthesis of proteins. J. molec. Biol.3 (1961) 318–356.
Johnson, A. D., and Herskowitz, I., A repressor (MATα2 product) and its operator control expression of a set of cell-type specific genes in yeast. Cell42 (1985) 237–247.
Johnston, M., A model fungal gene regulatory mechanism: the GAL genes ofSaccharomyces cerevisiae. Microbiol. Rev.51 (1987) 458–476.
Johnston, M., Genetic evidence that zinc is an essential co-factor in the DNA binding domain of GAL4 protein. Nature328 (1987) 353–355.
Johnston, M., and Dover, J., Mutational analysis of the Gal4-encoded transcriptional activator protein ofSaccharomyces cerevisiae. Genetics120 (1988) 63–74.
Jones, R. H., Moreno, S., Nurse, P., and Jones, N. C., Expression of the SV40 promoter in fission yeast: identification and characterization of an AP-1-like factor. Cell53 (1988) 659–667.
Jones, N. C., Rigby, P. W. J., and Ziff, E. B., Trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev.2 (1988) 267–281.
Kakidani, H., and Ptashne, M., Gal4 activates gene expression in mammalian cells. Cell52 (1988) 161–167.
Kaufer, N. F., Simanis, V., and Nurse, P., Fission yeastSchizosaccharomyces pombe correctly exises a mammalian RNA transcript intervening sequence. Nature318 (1985) 78–80.
Klein-Hitpaß, Schorpp, M., Wagner, U., and Ryffel, G. U., An estrogen-responsive element derived from the 5′ flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell46 (1986) 1053–1061.
Klug, A., and Rhodes, D., ‘Zinc fingers’: a novel protein motif for nucleic acid recognition. Trends biochem. Sci.12 (1987) 464–469.
Kornuc, M., Altman, R., Harrich, D., Garcia, H., Chao, J., Kayne, P., and Gaynor, R., Adenovirus transcriptional regulatory regions are conserved in mammalian cells andSaccharomyces cerevisiae. Molec. cell. Biol.8 (1988) 3717–3725.
Lambert, P. F., Dostatni, N., McBride, A. A., Yaniv, M., Howley, P. M., and Arcangioli, B., Functional analysis of the papilloma virus E2 trans-activator inSaccharomyces cerevisiae. Genes Dev.3 (1989) 38–48.
Landschulz, W. F., Johnson, P. F., and McKnight, S. L., The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science240 (1988) 1759–1764.
Landschultz, W. H., Johnson, P. F., Adoshi, E.Y., Graves, B. J. and McKnight, S. L., Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev.2 (1988) 786–800.
Lech, K., Anderson, K., and Brent, R., DNA-bound Fos proteins activate transcription in yeast. Cell52 (1988) 179–184.
Lee, W., Mitchell, P., and Tjian, R., Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell49 (1987) 741–752.
Leu, N. F., and Kornberg, R. D., Accurate initiation at RNA polymerase II promoter in extracts fromSaccharomyces cerevisiae. Proc. natl Acad. Sci. USA84 (1987) 8839–8843.
Levine, M., and Hoey, T., Homeobox proteins as sequence-specific transcription factors. Cell55 (1988) 537–540.
Lindquist, S., The heat-shock response. A. Rev. Biochem.55 (1986) 1151–1191.
Ma, J., Prizbilla, E., Hu, J., Bogorad, L., and Ptashne, M., Yeast activators stimulate plant gene expression. Nature334 (1988) 631–633.
Ma, L. and Ptashne, M., A new class of yeast transcriptional activators. Cell51 (1987) 113–119.
Ma, J., and Ptashne, M., Deletion analysis of Gal4 defines two transcription activating segments. Cell48 (1987) 847–853.
McBride, A. A., Schelegel, R., and Howley, P. M., The carboxy-terminal domain shared by the bovine papillomavirus E2 transactivator and repressor proteins contains a specific DNA binding activity. EMBO J.7 (1988) 533–539.
McClure, W. R., Mechanism and control of transcription initiation in prokaryotes. A. Rev. Biochem.54 (1985) 171–204.
McKnight, S., and Tjian, R., Transcriptional selectivity of viral genes in mammalian cells. Cell46 (1986) 795–805.
Miesfeld, R., Godowski, P. J., Maler, B. A., and Yamamoto, K. R., Glucocorticoid receptor mutants that define a small region sufficient for enhancer activation. Science236 (1987) 423–427.
Metzger, D., White, J. H., and Chambon, P., The human oestrogen receptor functions in yeast. Nature334 (1988) 31–36.
Miller, J., McLachland, A. D., and Klug, A., Repetitive zinc-binding domains in the protein transcription factor IIIA fromXenopus oocytes. EMBO J.4 (1985) 1609–1614.
Nonet, M., Sweetster, R. A., and Young, R. A., Functional redundacy and structural polymorphisms in the large subunit of RNA polymerase II. Cell50 (1987) 909–915.
Olesen, J., Hahn, S., and Guarente, L., Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell51 (1987) 953–961.
Pabo, C. O., and Sauer, R. T., Protein-DNA recognition. A. Rev. Biochem.53 (1984) 293–321.
Parker, C. S., and Topol, J., ADrosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp70 gene. Cell37 (1984) 273–283.
Payvar, F., DeFranco, D., Firestone, G. L., Edgar, B., Wrange, O., Okret, S., Gustafsson, J.-A., and Yamamoto, K. R., Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell35 (1983) 381–392.
Pelham, H., Activation of heat-shock gene in eukaryotes. Trends Genet.1 (1985) 31–35.
Pelham, H. R. B., A regulatory upstream promoter element in theDrosophila hsp70 heat-shock gene. Cell30 (1982) 517–528.
Pelham, H. R. B., and Bienz, M., A synthetic heat-shock promoter element confers heat-inducibility on the herpes simplex virus thymidine kinase gene. EMBO J.1 (1982) 1473–1477.
Picard, D., Salser, S. J., and Yamamoto, K. R., A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell54 (1988) 1073–1080.
Ponta, H., Kennedy, N., Skroch, P., Hynes, N. E., and Groner, B., Hormonal response region in the mouse mammary tumor virus long terminal repeat can be dissociated from the proviral promoter and has enhancer properties. Proc. natl Acad. Sci. USA82 (1985) 1020–1024.
Popham, D. L., Szeto, D., Keener, J., and Kustu, S., Function of bacterial activator protein that binds to transcriptional enhancers. Science243 (1989) 629–635.
Pratt, W. B., Jolly, D. J., Pratt, D. V., Hollenberg, S. M., Giguere, V., Cadepond, F. M., Schweizer-Groyer, G., Catelli, M.-G., Evans, R. M., and Baulieu, E.-E., A region in the steroid binding domain determines formation of the non-DNA binding, 9S glucocorticoid receptor complex. J. biol. Chem.263 (1988) 267–273.
Ptashne, M., A Genetic Switch: Gene Control and Phage λ. Cell Press & Blackwell Scientific Publications. Cambridge, Massachusetts 1986.
Ptashne, M., Gene regulation by proteins acting nearby and at a distance. Nature322 (1986) 697–701.
Ptashne, M., How eukaryotic transcriptional activators work. Nature335 (1988) 683–689.
Ptashne, M., How gene activators work. Sci. Am.260 (1989) 41–47.
Reinberg, D., and Roeder, R. G., Factors involved in specific transcription by mammalian RNA polymerase II. J. biol. Chem.262 (1987) 3310–3321.
Reitzer, L. J., and Magasanik, B., Transcription ofglnA inE. coli is stimulated by activator bound to sites far from the promoter. Cell45 (1986) 785–792.
Russell, P. R., Evolutionary divergence of the mRNA transcription initiation mechanism in yeast. Nature301 (1983) 167–169.
Sadowski, I., Ma, J., Triezenberg, S., and Ptashne, M., GAL4-VP16 is an unusually potent transcriptional activator. Nature335 (1988) 563–564.
Saltzman, A. G., and Weinmann, R., Promoter specificity and modulation of RNA polymerase II transcription. FASEB J.3 (1989) 1723–1733.
Samuels, M., Fire, A., and Sharp, P. A., Separation and characterization of factors mediating accurate transcription by RNA polymerase II. J. biol. Chem.267 (1982) 14 419–14 427.
Sauer, R. T., Smith, D. L., and Johnson, A. D., Flexibility of the yeast α-2 repressor enables it to occupy the ends of its operator, leaving the center free. Genes Dev.2 (1988) 807–816.
Scheidereit, C., Geisse, S., Westphal, H. M., and Beato, M., The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus. Nature304 (1983) 749–752.
Schena, M., and Yamamoto, K. R., Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science241 (1988) 965–967.
Scott, M. P., and O'Farrell, P. H., Spatial programming of gene expression in earlyDrosophila embryogenesis. A. Rev. Cell Biol.2 (1986) 49–80.
Sentenac, A., Eukaryotic RNA polymerases. CRC Crit. Rev. Biochem.18 (1985) 31–90.
Sigler, P. B., Acid blobs and negative noodles. Nature333 (1988) 210–212.
Soeller, W. C., Poole, S. J., and Kornberg, T., In vitro transcription of theDrosophila engrailed gene. Genes Dev.2 (1988) 68–81.
Sorger, P. K., and Pelham, H. R. B., Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell54 (1988) 855–864.
Sorger, P. K., and Pelham, H. R. B., Purification and characterization of a heat-shock element binding protein from yeast. EMBO J.6 (1987) 3035–3041.
Struhl, K., The Jun oncoprotein, a vertebrate transcription factor, activates transcription in yeast. Nature332 (1988) 649–651.
Struhl, K., The DNA-binding domains of the Jun oncoprotein and the yeast GCN4 transcriptional activator protein are functionally homologous. Cell50 (1987) 841–846.
Struhl, K., Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell49 (1987) 295–297.
Struhl, K., Helix-turn-helix, zinc-finger, and leucine zipper motifs for eukaryotic transcriptional regulatory proteins. TIBS14 (1989) 137–140.
Triezenberg, S. J., Kingsbury, R. C., and McKnight, S. L., Functional dissection of VP-16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev.2 (1988) 718–729.
Vogt, P. K., Bos, T. J., and Doolittle, R. F., Homology between the DNA-binding domain of the GCN4 regulatory protein of yeast and the carboxyl-terminal region of a protein coded for by the oncogene jun. Proc. natl Acad. Sci. USA84 (1987) 3316–3319.
Watts, F., Castle, C., and Beggs, J., Aberrant splicing ofDrosophila alcohol dehydrogenase transcripts inSaccharomyces cerevisiae. EMBO J.2 (1983) 2085–2091.
Webster, N., Jin, J.R., Green, S., Hollis, M., and Chambon, P., The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the Gal4 trans-activator. Cell52 (1988) 169–178.
Wiederrecht, G., Seto, D., and Parker, C. S., Isolation of the gene encoding theS. cerevisiae heat shock transcription factor. Cell54 (1988) 841–853.
Wiederrecht, G., Shuey, D. J., Kibbe, W. A., and Parker, C. S., TheSaccharomyces andDrosophila heat shock transcription factors are identical in size and DNa binding properties. Cell48 (1987) 507–515.
Weinberger, C., Hollenberg, S. M., Rosenfeld, M. G., and Evans, R. M., Domain structure of human glucocorticoid receptor and its relationship of the v-erbA oncogene product. Nature318 (1985) 670–672.
Yamamoto, K. R., Stampfer, M. R., and Tompkins, G. M., Receptors from glucocorticoid-sensitive lymphoma cells and two classes of insensitive clones: physical and DNA-binding properties. Proc. natl Acad. Sci. USA71 (1974) 3901–3905.
Yamamoto, K. R., Steroid receptor regulated transcription of specific genes and gene networks. A. Rev. Genet.19 (1985) 209–252.
Yamamoto, K. R., A conceptual view of transcriptional regulation. Am. Zool.29 (1989) 537–547.
Yura, T., and Ishihama, A., Genetics of bacterial RNA polymerases. A. Rev. Genet.13 (1979) 59–97.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Schena, M. The evolutionary conservation of eukaryotic gene transcription. Experientia 45, 972–983 (1989). https://doi.org/10.1007/BF01953055
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF01953055