Abstract
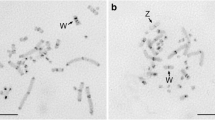
In studies on the highly repetitive DNA sequences of the flesh flySarcophaga bullata, a 279 bp tandem repeat was cloned and sequenced. A 17 bp stretch within the clone was identical to a motif repeated five times in the satellite DNA of the Bermuda land crab. Southern DNA blotting showed the tandem repeat had a high degree of conservation of MboI sites, but had divergence for EcoRI sites; thus, all repeat units were not identical. The cloned DNA localized to the quinacrine-bright centromeric heterochromatin of the C and E autosomes and to sites on the chromosomal arms. In cases of asynapsis of homologs, the probe localized to euchromatic sites on both homologs or sometimes only on one homolog. The probe also localized near, to, or at a major developmental puff (B9). We conclude that blocks of this short interspersed repetitive DNA occur throughout theSarcophaga genome in both heterochromatin and euchromatin, and also that the variable position of these sequences suggests they possess a degree of instability.
Similar content being viewed by others
References
Bonnewell V, Fowler RF, Skinner DM (1983) An inverted repeat borders a fivefold amplification in satellite DNA. Science 221:862–865
Bultmann H, Clever U (1969) Chromosomal control of foot pad development inSarcophaga bullata. I. The puffing pattern. Chromosoma 28:120–135
Bultmann H, Mezzanotte R (1987) Characterization and origin of extrachromosomal DNA granules inSarcophaga bullata. J Cell Sci 88:327–334
Crain WR, Davidson EH, Britten RJ (1976) Contrasting patterns of DNA sequence arrangement inApis mellifera (honeybee) andMusca domestica (house fly). Chromosoma 59:1–12
Fowler RF, Bonnewell V, Spann MS, Skinner DM (1985) Sequences of three closely related variants of a complex satellite DNA diverge at specific domains. J Biol Chem 260:8964–8972
Kaul D, Gaur P, Agrawal UR, Tewari RR (1989a) Characterization of Parasarcophaga heterochromatin. I. Mitotic chromosomes. Chromosoma 98:49–55
Kaul D, Agrawal UR, Tewari RR (1989b) Characterization of Parasarcophaga heterochromatin. II. Polytene chromosomes. Chromosoma 98:56–63
Leary JJ, Brigati DJ, Ward DC (1983) Rapid and sensitive colorimetric method for visualizing biotin-labeled DNA probes hybridized to DNA or RNA immobilized on nitrocellulose: Bioblots. Proc Natl Acad Sci USA 80:4045–4049
Lifschytz E, Hareven D (1982) Heterochromatin markers: A search for heterochromatin specific middle repetitive sequences inDrosophila. Chromosoma 86:429–442
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Pardue ML, Dawid IB (1981) Chromosomal locations of two DNA segments that flank ribosomal insertion-like sequences inDrosophila: flanking sequences are mobile elements. Chromosoma 83:29–43
Roberts B, Whitten JM, Gilbert LI (1976) Patterns of incorporation of tritiated thymidine in the dorsal polytene foot-pad nuclei ofSarcophaga bullata (Sarcophagidae: Diptera). Chromosoma 54:127–140
Samols DR (1976) Characterization of the genome of the flesh flySarcophaga bullata. Ph.D. Thesis. The University of Chicago, Chicago
Samols D, Swift H (1979a) Genomic organization in the flesh flySarcophaga bullata. Chromosoma 75:129–143
Samols D, Swift H (1979b) Characterization of extrachromosomal DNA in the flesh flySarcophaga bullata. Chromosoma 75:145–159
Specht CA, DiRusso CC, Novotny CP, Ullrich RC (1982) A method for extracting high-molecular-weight deoxyribonucleic acid from fungi. Anal Biochem 119:158–163
Truett MA, Jones RS, Potter SS (1981) Unusual structure of the FB family of transposable elements inDrosophila. Cell 24:753–763
Whitten JM (1969) Coordinated development in the foot pad of the flySarcophaga bullata during metamorphosis: changing puffing patterns of the giant cell chromosomes. Chromosoma 26:215–244
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Hershfield, B., Swift, H. Characterization of a tandemly repeated DNA from the fleshflySarcophaga bullata . Chromosoma 99, 125–130 (1990). https://doi.org/10.1007/BF01735328
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01735328