Summary
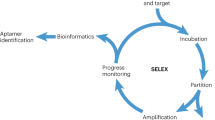
In vitro selection, or SELEX, has been used both to characterize the interaction of natural nucleic acids with proteins and to generate novel nucleic acid-binding species, or aptamers. Although numerous reports have demonstrated the power of the technique, they have not expanded on the methodologies that can be used for selection. This review focuses on the considerations and problems involved in selecting protein-binding aptamers from a random-sequence RNA pool. As an illustration, we describe two approaches to selecting aptamers to a particular target, the HTLV-I Rex protein. In the first, complete randomization is used to find an artificial, high-affinity RNA binding site. In the second, the contributions of individual nucleotides and/or base pairs to the natural Rex-binding element are determined by mutating the wild-type sequence and selecting active binding variants.
Similar content being viewed by others
References
Conrad, R.C., Giver, L., Tian, Y. and Ellington, A.D.,In vitro selection of nucleic acid aptamers that bind proteins, Methods Enzymol., (1995) in press.
Ellington, A. and Conrad, R.,Aptamers as potential nucleic acid pharmaceuticals, In Biotechnology Annual Review, Vol. 1, Elsevier, Amsterdam, 1995, in press.
Gold, L.,Oligonudeotides as research, diagnostic, and therapeutic agents, J. Biol. Chem., 270 (1995) 13581–13584.
Gold, L., Polisky, B., Uhlenbeck, O. and Yarus, M.,Diversity of oligonudeotide functions, Annu. Rev. Biochem., 64 (1995) 763–797.
Oliphant, A.R., Brandi, C.J. and Strahl, K.,Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: Analysis of GCN4 protein, Mol. Cell. Biol., 9 (1989) 2944–2949.
Tuerk, C. and Gold, L.,Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase, Science, 249 (1990) 505–510.
Schneider, D., Tuerk, C. and Gold, L.,Selection of high affinity RNA ligands to the bacteriophage R17 coat protein, J. Mol. Biol., 228 (1992) 862–869.
Schneider, D., Gold, L. and Platt, T.,Selective enrichment of RNA species for tight binding to Escherichia coli rho factor, FASEB J., 7 (1993) 201–207.
Tsai, D.E., Harper, D.S. and Keene, J.D.,U1-snRNP-A protein selects a ten nucleotide consensus sequence from a degenerate RNA pool presented in various structural contexts, Nucleic Acids Res., 19 (1991) 4931–4936.
Levine, T.D., Gao, F., King, P.H., Andrews, L.G. and Keene, J.D.,Hel-N1: An autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs, Mol. Cell. Biol., 13 (1993) 3494–3504.
Tsai, D.E., Kenan, D.J. and Keene, J.D.,In vitro selection of an RNA epitope immunologically cross-reactive with a peptide, Proc. Natl. Acad. Sci. USA, 89 (1992) 8864–8868.
Tsai, D.E. and Keene, J.D.,In vitro selection of RNA epitopes using autoimmune patient serum, J. Immunol., 150 (1993) 1137–1145.
Jellinek, D., Lynott, C.K. and Janjic, D.B.R.N,High-affinity RNA ligands to basic fibroblast growth factor inhibit receptor binding, Proc. Natl. Acad. Sci. USA, 90 (1993) 11227–11231.
Conrad, R., Keranen, L.M., Ellington, A.D. and Newton, A.C.,Isozyme-specific inhibition of protein kinase C by RNA aptamers, J. Biol. Chem., 269 (1994) 32051–32054.
Tian, Y., Adya, N., Wagner, S., Giam, C.-z., Green, M.R. and Ellington, A.D.,Dissecting protein:protein interactions between transcription factors with an RNA aptamer, RNA, 1 (1995) 317–326.
Bock, L.C., Griffin, L.C., Latham, J.A., Vermass, E.H. and Toole, J.J.,Selection of single-stranded DNA molecules that bind and inhibit human thrombin, Nature, 355 (1992) 564–566.
Padmanabhan, K., Padmanabhan, K.P., Ferrera, J.D., Sadler, J.E. and Tulinsky, A.,The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer, J. Biol. Chem., 268 (1993) 17651–17654.
Kubik, M.F., Stephens, A.W., Schneider, D., Marlar, R.A. and Tasset, D.,High-affinity RNA ligands to human α-thrombin, Nucleic Acids Res., 22 (1994) 2619–2626.
Bartel, D.P., Zapp, M.L., Green, M.R. and Szostak, J.W.,HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA, Cell, 67 (1991) 529–536.
Giver, L., Bartel, D., Zapp, M., Pawul, A., Green, M. and Ellington, A.D.,Selective optimization of the Rev-binding element of HIV-1, Nucleic Acids Res., 21 (1993) 5509–5516.
Tuerk, C. and MacDougal-Waugh, S.,In vitro evolution of functional nucleic acids: High-affinity ligands of HIV-1 proteins, Gene, 137 (1993) 33–39.
Tuerk, C., MacDougal, S. and Gold, L.,RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase, Proc. Natl. Acad. Sci. USA, 89 (1992) 6988–6992.
Chen, H. and Gold, L.,Selection of high-affinity RNA ligands to reverse transcriptase: Inhibition of cDNA synthesis and RNase H activity, Biochemistry, 33 (1994) 8746–8756.
Jenison, R.D., Gill, S.C., Pardi, A. and Polisky, B.,High-resolution molecular discrimination by RNA, Science, 263 (1994) 1425–1429.
Paborsky, L.R., McCurdy, S.N., Griffin, L.C., Toole, J.J. and Leung, L.L.K.,The single-stranded DNA aptamer-binding site of human thrombin, J. Biol. Chem., 268 (1993) 20808–20811.
Peterson, E.T., Blank, J., Spronz, M. and Uhlenbeck, O.C.,Selection for active E. coli tRNA phe variants from a randomized library using two proteins, EMBO J., 12 (1993) 2959–2967.
Peterson, E.T., Pan, T., Coleman, J. and Uhlenbeck, O.C.,In vitro selection of small RNAs that bind to Escherichia coli phenylalanyltRNA synthetase, J. Mol. Biol., 242 (1994) 186–192.
Leclerc, F., Cedegren, R. and Ellington, A.,A three-dimensional model of the Rev-binding element of HIV-1 derived from analyses of aptamers, Nature Struct. Biol., 1 (1994) 293–300.
Kramer, F.R., Mills, D.R., Cole, P.E., Nishihara, T. and Spiegelman, S.,Evolution of in vitro sequence and phenotype of a mutant RNA resistant to ethidium bromide, J. Mol. Biol., 4 (1974) 427–429.
Orgel, L.E.,Selection in vitro, Proc. R. Soc. London, 205 (1979) 435–442.
Abmayr, S.B. and Workman, J.L.,Mobility shift DNA-binding assay using gel electrophoresis, In Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A. and Struhl, K. (Eds.) Current Protocols in Molecular Biology, Vol. 2, Wiley, New York, NY, 1988, pp. 12.2.1–12.2.6.
Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G. and Erlich, H.,Specific enzymatic amplification of DNA in vitro: The polymerase chain reaction, Cold Spring Harbor Symp. Quant. Biol., 51 (1986) 263–273.
Guatelli, J.C., Whitfield, K.M., Kwoh, D.Y., Barringer, K.J., Richman, D.D. and Gingeras, T.R.,Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modelled after retroviral replication, Proc. Natl. Acad. Sci. USA, 87 (1990) 1874–1878.
Kwoh, D.Y., Davis, G.R., Whitfield, K.M., Chapelle, H.L., DeMichele, L.J. and Gingeras, T.R.,Transcription-based amplification system and detection of amplified human immunodeficiency virus type 1 with a bead-based sandwich hybridization format, Proc. Natl. Acad. Sci. USA, 86 (1989) 1173–1177.
Melton, D.A., Krieg, P.A., Rebagliati, M.R., Maniatis, T., Zinn, K. and Green, M.R.,Efficient in vitro synthesis of active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter, Nucleic Acids Res., 12 (1984) 7035–7056.
Milligan, J.F., Groebe, D.R., Witherell, G.W. and Uhlenbeck, O.C.,Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates, Nucleic Acids Res., 15 (1987) 8783–8798.
Lato, S.M., Boles, A.R. and Ellington, A.D.,In vitro selection of RNA lectins: Using combinatorial chemistry to interpret ribozyme evolution, Chem. Biol., 2 (1995) 291–303.
Lorsch, J.R. and Szostak, J.W.,In vitro selection of RNA aptamers specific for cyanocobalamin, Biochemistry, 33 (1994) 973–982.
Irvine, D., Tuerk, C. and Gold, L.,SELEXION systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis, J. Mol. Biol., 222 (1991) 739–761.
Ellington, A. and Green, R.,Synthesis of oligonucleotides, In Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A. and Struhl, K. (Eds.) Current Protocols in Molecular Biology, Vol. 1, Wiley, New York, NY, 1988, pp. 2.11.1–2.11.18.
Dicker, A.P., Volkenandt, M. and Bertino, J.R.,Manual and automated direct sequencing of product generated by the polymerase chain reaction, In White, B.A. (Ed.) Methods in Molecular Biology, Vol. 15: PCR Protocols: Current Methods and Applications, Humana Press, Totowa, NJ, 1993, pp. 143–152.
Silveira, M.H. and Orgel, L.E.,PCR with detachable primers, Nucleic Acids Res., 23 (1995) 1083–1084.
Latham, J.A., Johnson, R. and Toole, J.J.,The application of a modified nucleotide in aptamer selection: Novel thrombin aptamers containing 5-(1-pentynyl)-2′-deoxyuridine, Nucleic Acids Res., 22 (1994) 2817–2822.
Aurup, H., Williams, D.M. and Eckstein, F.,2′-fluoro- and 2′-amino-2′-deoxynucleoside 5′-triphosphates as substrates for T7 RNA polymerase, Biochemistry, 31 (1992) 9636–9641.
Lin, Y., Qiu, Q., Gill, S.C. and Jayasena, S.D.,Modified RNA sequence pools for in vitro selection, Nucleic Acids Res., 22 (1994) 5229–5234.
Ruffner, D.E. and Uhlenbeck, O.C.,Thiophosphate interference experiments locate phosphates important for the hammerhead RNA self-cleavage reaction, Nucleic Acids Res., 18 (1990) 6025–6029.
Crameri, A. and Stemmer, W.P.C., 1020-fold aptamer library amplification without gel purification, Nucleic Acids Res., 21 (1993) 4410.
Ahmed, Y.F., Hanley, S.M., Malim, M.H., Cullen, B.R. and Greene, W.C.,Structure-function analyses of the HTLV-I Rex and HIV-1 Rev RNA response elements: Insights into the mechanism of Rex and Rev action, Genes Dev., 4 (1990) 1014–1022.
Bogerd, H.P., Huckaby, G.L., Ahmed, Y.F., Hanly, S.M. and Greene, W.C.,The type I human T-cell leukemia virus (HTLV-I) Rex trans-activator binds directly to the HTLV-I Rex and the type 1 human immunodeficiency virus Rev RNA response elements, Proc. Natl. Acad. Sci. USA, 88 (1991) 5704–5708.
Baskerville, S., Zapp, M. and Ellington, A.D.,High-resolution mapping of the HTLV-I Rex binding element by in vitro selection, J. Virology, (1995) in press.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Conrad, R.C., Baskerville, S. & Ellington, A.D. In vitro selection methodologies to probe RNA function and structure. Mol Divers 1, 69–78 (1995). https://doi.org/10.1007/BF01715810
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01715810