Abstract
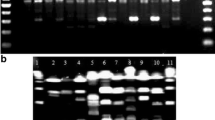
Although several typing methods have been described for Shiga-like toxin-producingEscherichia coli O157, the methods are somewhat cumbersome. Using 22 isolates ofEscherichia coli O157 and five otherEscherichia coli isolates, two primers (M13 core sequence and 970-11) were found to give excellent differentiation between isolates using random amplified polymorphic DNA (RAPD). Using only the presence or absence of variable bands, a matrix of 20 variable characters was identified. From these characters, similarity coefficients were calculated and a phenogram constructed. All of theEscherichia coli O157 isolates were easily distinguished from the non-O157Escherichia coli isolates. Using a 95% similarity cutoff, we found 13 RAPD types among the 22Escherichia coli O157 isolates. Isolates thought to be identical by toxin and phage typing as well as by epidemiological association were distinguished, and others thought to be distinct by lack of epidemiological association were identical. RAPD using M13 and 970-11 primers is a potentially useful typing tool forEscherichia coli isolates of serotype O157 and possibly otherEscherichia coli isolates.
Similar content being viewed by others
References
Griffin PM, Tauxe RV: The epidemiology of infections caused byEscherichia coli O157:H7, other enterohemorrhagicE. coli, and the associated hemolytic uremic syndrome. Epidemiologic Reviews 1991, 13: 60–98.
Karmali MA: Infection by verocytotoxin-producingEscherichia coli. Clinical Microbiology Reviews 1989, 2: 15–38.
Whittam TS, Wolfe ML, Wachsmuth K, Orskov F, Orskov I, Wilson RA: Clonal relationships amongEscherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infection and Immunity 1993, 61: 1619–1629.
Whittam TS, Wachsmuth IK, Wilson RA: Genetic evidence of clonal descent ofEscherichia coli O157:H7 associated with hemorrhagic colitis and hemolytic uremic syndrome. Journal of Infectious Diseases 1988, 157: 1124–1133.
Abbott SL, Hanson DF, Felland TD, Connell S, Shum AH, Janda JM:Escherichia coli O157:H7 generates a unique biochemical profile on microscan conventional gram-negative identification panels. Journal of Clinical Microbiology 1994, 32: 823–824.
Ostroff SM, Tarr PI, Neill MA, Lewis JH, Hargrett-Beann N, Kobayashi JM: Toxin genotypes and plasmid profiles as determinants of systemic sequelae inEscherichia coli O157:H7 infections. Journal of Infectious Diseases 1989, 160: 994–998.
Barrett TJ, Lior H, Green JH, Khakhria R, Wells JG, Bell BP, Greene KD, Lewis J, Griffin PM: Laboratory investigation of a multistate food-borne outbreak ofEscherichia coli O157-H7 by using pulsed-field gel electrophoresis and phage typing. Journal of Clinical Microbiology 1994, 32:3013–3017.
Frost JA, Cheasty T, Thomas A, Rowe B: Phage typing of Vero cytotoxin-producingEscherichia coli O157:H7 isolated in the United Kingdom: 1989–91. Epidemiology and Infection 1993, 110: 469–475.
Bohm H, Karch H: DNA fingerprinting ofEscherichia coli O157:H7 strains by pulsed-field gel electrophoresis. Journal of Clinical Microbiology 1992, 30: 2169–2172.
Samadpour M, Grimm LM, Desai B, Alfi D, Ongerth JE, Tarr PI: Molecular epidemiology ofEscherichia coli O157:H7 strains by bacteriophage λ-restriction fragment length polymorphism analysis: application to a multistate foodborne outbreak and a day-care centre. Journal of Clinical Microbiology 1993, 31: 3179–3182.
Dorn CR, Angrick EJ: Serotype O157:H7Escherichia coli from bovine and meat sources. Journal of Clinical Microbiology 1991, 29: 1225–1231.
Chatelut M, Dournes JL, Chabanon G, Marty N: Epidemiological typing ofStenotrophomonas (Xanthomonas) maltophilia by PCR. Journal of Clinical Microbiology 1995, 33:912–914.
Corney BG, Colley J, Djordjevic SP, Whittington R, Graham GC: Rapid identification of someLeptospira isolates from cattle by random amplified polymorphic DNA fingerprinting. Journal of Clinical Microbiology 1993, 31: 2927–2932.
Seppala H, He Q, Osterblad M, Huovinen P: Typing of group A streptococci by random amplified polymorphic DNA analysis. Journal of Clinical Microbiology 1994, 32: 1945–1948.
Wang G, Whittam TS, Berg CM, Berg DE: Rapid (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Research 1993, 21: 5930–5933.
Law D, Hamour AA, Acheson DWK, Panigrahi H, Ganguli LA, Denning DW: Diagnosis of infection with Shiga-like toxin producingEscherichia coli by use of enzyme linked immunosorbent assays for Shiga-like toxins on cultured stool samples. Journal of Medical Microbiology 1994, 40:241–245.
Cebula TA, Payne WL, Feng P: Simultaneous identification of strains ofEscherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. Journal of Clinical Microbiology 1995, 33: 248–250.
Aufauvre-Brown A, Cohen J, Holden DW: Use of randomly amplified polymorphic DNA markers to distinguish isolates ofAspergillus fumigatus. Journal of Clinical Microbiology 1992, 30: 2991–2993.
van Belkum A, Melchers WJ, de Pauw BE: Genotypic characterisation of sequentialCandida albicans isolates from fluconazole-treated neutropenic patients. Journal of Infectious Diseases 1994, 169: 1062–1070.
Schonian G, Meusel O, Tietz HJ, Meyer W, Graser Y, Tausch I, Presber W, Mitchell TG: Identification of clinical strains ofCandida albicans by DNA fingerprinting with the polymerase chain reaction. Mycoses 1993, 36: 171–179.
Menard C, Mouton C: Randomly amplified polymorphic DNA analysis confirms the biotyping scheme ofPorphyromonas gingivalis. Research in Microbiology 1993, 144:445–455.
Paros M, Tarr PI, Kim H, Besser TE, Hancock DD: A comparison of human and bovineEscherichia coli O157:H7 isolates by toxin genotype, plasmid profile, and bacteriophage λ-restriction fragment length polymorphism profile. Journal of Infectious Diseases 1993, 168: 1300–1303.
Bandi C, La Rosa G, Bardin MG, Damiani G, Cominci S, Tasciotti L, Pozio E: Random amplified polymorphic DNA fingerprints of the eight taxa ofTrichenella and their comparison with allozyme analysis. Parasitology 1995, 110:401–407.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Birch, M., Denning, D.W. & Law, D. Rapid genotyping ofEscherichia coli O157 isolates by random amplification of polymorphic DNA. Eur. J. Clin. Microbiol. Infect. Dis. 15, 297–302 (1996). https://doi.org/10.1007/BF01695661
Issue Date:
DOI: https://doi.org/10.1007/BF01695661