Abstract
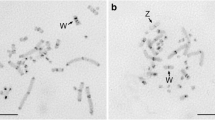
A highly repetitive DNA clone, designated Rt-Pst3, was isolated from thePstI digest of Canadian woodland caribou (Rangifer tarandus caribou; 2n = 70) genomic DNA. It was found to be a 991 bp monomer of a tandemly repeated DNA sequence comprising about 5.7% of the genome and localized to the centromeric regions of all caribou acrocentric autosomes. Southern blot analyses revealed that this caribou satellite DNA sequence was well conserved in the genomes of five other deer species studied.In situ hybridization studies revealed Rt-Pst3-homologous DNA sequences in the centromeric regions of white-tailed deer chromosomes and Asian muntjac chromosomes, as well as at several interstitial chromosome regions in Indian muntjac chromosomes. Comparisons of the Rt-Pst3 DNA sequence to previously identified centromeric satellite DNA fragments from three other deer species revealed considerable DNA sequence similarity. The first ca. 800 bp of the Rt-Pst3 clone was found to share 73.8% similarity to theCCsatI clone of the European roe deer, 64.7% sequence similarity to the C5 DNA clone of the Chinese muntjac, and 64.8% and 65.6% sequence similarity to the 1A and B1 clones of the Indian muntjac, respectively. Moreover, the last 191 bp of the Rt-Pst3 clone was found to share about 60% DNA sequence similarity to the first 191 bp of the same clone. Amplification of one originalca. 800 bp monomer unit, along with the first 191 bp of the following juxtaposed monomer unit could have resulted in the tandemly repeated, 991 bp monomer unit now seen in the caribou genome. It is postulated that the centromeric satellite DNA found in other deer species, having repeat lengths greater than 800 bp, could also have evolved in a similar manner from a more ancestral monomeric unit ofca. 800 bp.
Similar content being viewed by others
References
Armud J, Nes N (1966) The chromosomes of the Roe (Capreolus capreolus).Hereditas 56: 217–220.
Baccus R, Ryman N, Smith MH, Reuterwall C, Cameron D (1983) Genetic variability and differentiation of large grazing mammals.J Mammal 64: 109–120.
Banfield AWF (1961) A revision of the reindeer and caribou, genusRangifer.Bull Natl Mus Can 177 (Biol ser66 pp 1–137.
Bogenberger JM, Neumaier PS, Fittler F (1985) The Muntjak satellite IA sequence is composed of 31-bp-pair internal repeats that are highly homologous to the 31-base-pair subrepeats of the bovine satellite 1.715.Eur J Biochem 148: 55–59.
Bogenberger JM, Neitzel H, Fittler F (1987) A highly repetitive DNA component common to all cervidae: its organization and chromosomal distribution during evolution.Chromosoma 95: 154–161.
Davies LG, Dibner MD, Battey JF (1986) DNA preparation from cultured cells and tissue. In: Davies LG, Dibner MD, Battey JF, eds.Basic Methods in Molecular Biology, New York: Elsevier, pp. 47–50.
Durfy SJ, Willard HF (1989) Patterns of intra- and interarray sequence variation in alpha satellite from the human X chromosome: evidence for short-range homogenization of tandemly repeated DNA sequences.Genomics 5: 810–821.
Elder FFB, Hsu TC (1988) Tandem fusions in the evolution of mammalian chromosomes. In: Sandberg AA, ed.The Cytogenetics of Mammalian Autosomal Rearrangements. New York: AR Liss, pp. 481–506.
Emerson BC, Tate ML (1993) Genetic analysis of evolutionary relationships among deer (Subfamily Cervinae).J Heredity 84: 266–273.
Fontana F, Rubini M (1990) Chromosomal evolution in Cervidae.BioSystems 24: 157–174.
Fraccaro M, Gustavsson I, Hulten M, Lindsten J, Tiepolo L (1968) Chronology of DNA replication in the sex chromosomes of the reindeer (Rangifer tarandus L.).Cytogenetics 7: 196–211.
Gripenburg U (1984) Characterization of the karyotype of the reindeer (Rangifer tarandus); the distribution of the heterochromatin in the reindeer and the Scandinavian moose (Alces alces). In: Stranzinger G, ed.Proc 6th Eur Colloq Cytogenet Domest Anim Zurich. pp 68–79.
Hamilton MJ, Honeycutt RL, Baker RJ (1990) Intragenomic movement, sequence amplification, and concerted evolution inReithrodontomys: evidence fromin situ hybridization.Chromosoma 99: 321–329.
Henikoff S (1987) Unidirectional digestion with exonuclease III in DNA sequence analysis.Methods Enzymol 155: 156–165.
Horz W, Zachau HG (1977) Characterization of distinct segments in mouse satellite DNA by restriction nucleases.Eur J Biochem 73: 383–392.
Jeffreys AJ, Wilson V, Thein S (1985) Hypervariable minisatellite regions in human DNA.Nature 314: 67–73.
Kurten B (1971)The Age of Mammals. London: Weidenfield and Nicholson, pp. 250.
Lee C (1993) Tandemly repetitive DNA in the karyotypic and phylogenetic evolution of Cervidae species. MSc dissertation, Edmonton, Canada: University of Alberta.
Lee C, Sasi R, Lin CC (1993) Interstitial localization of telomeric DNA sequences in the Indian muntjac chromosomes: further evidence for tandem chromosome fusions in the karyotypic evolution of the Asian muntjacs.Cytogenet Cell Genet 63: 156–159.
Lima-de-Faria A (1980) How to produce a human with 3 chromosomes and 1000 primary genes.Hereditas 93: 47–73.
Lima-de-Faria A, Arnason U, Widegren Bet al. (1984) Conservation of repetitive DNA sequences in deer species studied by Southern blot transfer.J Mol Evol 20: 17–24.
Lima-de-Faria A, Arnason U, Widegren B, Isaksson M, Essen-Moller J, Jaworska H (1986) DNA cloning and hybridization in deer species supporting the chromosome field theory.Biosystems 19: 185–212.
Lin CC, Sasi R, Fan Y-S, Chen Z-Q (1991) New evidence for tandem chromosome fusions in the karyotypic evolution of Asian muntjacs.Chromosoma 101: 19–24.
Lin CC, Sasi R, Lee C, Fan YS, Court D (1993) Isolation and identification of a novel tandemly repeated DNA sequence in the centromeric region of human chromosome 8.Chromosoma 102: 333–339.
Maniatis T, Fritsch EF, Sambrook J (1982)Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
McDonnell MW, Simon MN, Studier FW (1977) Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels.J Mol Biol 110: 119–146.
Miyamoto MM, Kraus F, Ryder OA (1990) Phylogeny and evolution of antlered deer determined from mitochondrial DNA sequences.Proc Natl Acad Sci USA 87: 6127–6131.
Neitzel H (1982)Karyotypenevolution und deren Bedeutung fur den Speciationsprozess der Cerviden (Cervidae; Artiodactyla; Mammalia). Thesis, Freie Univ Berlin.
Neitzel H (1987) Chromosome evolution of cervidae: karyotypic and molecular aspects. In: Obe G, Basler A, eds.Cytogenetics. Berlin: Springer-Verlag, pp. 90–112.
Roed KH, Ferguson MAD, Crete M, Bergerud TA (1991) Genetic variation in transferrin as a predictor for differentiation and evolution of caribou from eastern Canada.Rangifer 11: 65–74.
Scherthan H (1991) Characterization of a tandem repetitive sequence cloned from the deerCapreolus capreolus and its chromosomal localisation in two muntjac species.Hereditas 115: 43–49.
Shi LM, Ye YY, Duan XS (1980) Comparative cytogenetic studies on the red muntjac, Chinese muntjac and their F1 hybrids.Cytogenet Cell Genet 26: 22–27.
Simpson CD (1984) Artiodactyls. In: Anderson S, Jones JK, eds.Orders and Families of Recent Mammals of the World. New York, Wiley. pp. 563–587.
Southern (1975) Long range periodicities in mouse satellite DNA.J Mol Biol 94: 51–69.
Steinmetz M, Stephan D, Lindahl FK (1986) Gene organization and recombinational hot spots in the murine histocompatibility complex.Cell 44: 895–904.
Yu LC, Lowensteiner D, Wong EFK, Sawada I, Mazrimas J, Schmid C (1986) Localization and characterization of recombinant DNA clones derived from the highly repetitive DNA sequences in the Indian muntjac cells: Their presence in the Chinese muntjac.Chromosoma 93: 521–528.
Wichman HA, Payne CT, Ryder OA, Hamilton MJ, Maltbie M, Baker RJ (1991) Genomic distribution of heterochromatic sequences in equids: Implications to rapid chromosomal evolution.J Heredity 82: 369–377.
Wurster DH, Benirschke K (1970) Indian muntjac,Muntiacus muntjak: a deer with a low diploid chromosome number.Science 168: 1364–1366.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Lee, C., Ritchie, D.B.C. & Lin, C.C. A tandemly repetitive, centromeric DNA sequence from the Canadian woodland caribou (Rangifer tarandus caribou): Its conservation and evolution in several deer species. Chromosome Res 2, 293–306 (1994). https://doi.org/10.1007/BF01552723
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01552723