Abstract
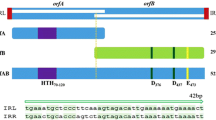
Bacillus thuringiensis is an entomopathogenic bacterium whose toxicity is due to the presence in the sporangia of δ-endotoxin crystals active against agricultural pests and vectors of human and animal diseases. Most of the genes coding for these toxin proteins are plasmid-borne and are generally structurally associated with insertion sequences (IS231, IS232, IS240, ISBT1 and ISBT2) and transposons (Tn4430 and Tn5401). Several of these mobile elements have been shown to be active and are believed to participate in the crystal gene mobility, thereby contributing to the variation of bacterial toxicity. Structural analysis of the iso-IS231 elements indicates that they are related to IS1151 fromClostridium perfringens and distantly related to IS4 and IS186 fromEscherichia coli. Like the other IS4 family members, they contain a conserved transposase-integrase motif found in other IS families and retroviruses. Moreover, functional data gathered from IS231 A inEscherichia coli indicate a non-replicative mode of transposition, with a marked preference for specific targets. Similar results were also obtained inBacillus subtilis andB. thuringiensis, and a working model for DNA-protein interactions at the target site is proposed.
Similar content being viewed by others
References
Adams, L. F., J. E. Visick & H. R. Whiteley, 1989. A 20-kilodalton protein is required for efficient production of theBacillus thuringiensis subsp.israelensis 27-kilodalton crystal protein inEscherichia coli. J. Bacteriol. 171: 521–530.
Ahmad, W., C. Nicholls & D. J. Ellar, 1989. Cloning and expression of an entomocidal protein gene fromBacillus thuringiensis galleriae toxic to both lepidoptera and diptera. FEMS Microbiol. Lett. 59: 197–202.
Aronson, A. I., 1993. The two faces ofBacillus thuringiensis: insecticidal proteins and post-exponential survival. Mol. Microbiol. 7: 489–496.
Baum, J., 1994. Tn5401: a new class II transposable element fromBacillus thuringiensis. J. Bacteriol. 176: 2835–2845.
Beese, L. S. & T. A. Steitz, 1991. Structural basis for the 3′–5′ exonuclease activity ofEscherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 10: 25–33.
Bender, J. & N. Kleckner, 1992. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the targetsite consensus sequence. Proc. Natl. Acad. Sci. USA 89: 7996–8000.
Bourgouin, C., A. Delécluse, J. Ribier, A. Klier & G. Rapoport, 1988. ABacillus thuringiensis gene encoding a 125-kilodalton larvicidal polypeptide is associated with inverted repeat sequences. J. Bacteriol. 170: 3575–3583.
Carlson, C. R., D. Caugant & A.-B. Kolstø, 1994. Genotypic diversity amongBacillus cereus andBacillus thuringiensis strains. Appl. Environ. Microbiol. 60: 1719–1725.
Carlson, C. R. & A.-B. Kolstø, 1993. A complete physical map of aBacillus thuringiensis chromosome. J. Bacteriol. 175: 1053–1060.
Chalker, D. L. & S. D. Sandmeyer, 1992. Ty3 integrates within the region of RNA polymerase III transcription initiation. Gene Dev. 6: 117–128.
Chong, P., I. Hui, T. Loo & S. Gilliam, 1985. Structural analysis of a new GC-specific insertion element IS186. FEBS Lett. 192: 47–52.
Craig, N., 1991. Tn7: a target site-specific transposon. Mol. Microbiol. 5: 2569–2573.
Daube, G., P. Simon & A. Kaeckenbeeck, 1993. IS1151, an IS-like element ofClostridium perfringens. Nucleic Acids Res. 21: 352.
Delécluse, A., C. Bourgouin, A. Klier & Rapoport, 1989. Nucleotide sequence and characterization of a new insertion element, IS240, fromBacillus thuringiensis israelensis. Plasmid 21: 71–78.
Delécluse, A., C. Bourgouin, G. Menou, D. Lereclus, A. Klier & G. Rapoport, 1990. IS240 associated with the cryIVA gene fromBacillus thuringiensis israelensis belongs to a family of Gram(+) and Gram(−) IS elements, pp. 181–190 in Genetics and Biotechnology of Bacilli, Vol. 3, edited by M. M. Zukowski, A. T. Ganesan and J. A. Hoch. Academic Press, San Diego, CA.
Delécluse, A., J.-F. Charles, A. Klier & G. Rapoport, 1991. Deletion by in vivo recombination shows that the 28-kilodalton cytolytic polypeptide fromBacillus thuringiensis subsp.israelensis is not essential for mosquitocidal activity. J. Bacteriol. 173: 3374–3381.
Derbyshire, V., N. D. F. Grindley & C. M. Joyce, 1991. The 3′–5′ exonuclease of DNA polymerase I ofEscherichia coli: contribution of each amino acid at the active site of the reaction. EMBO J. 10: 17–24.
Doak, T. G., F. P. Doerder, C. L. Jahn & G. Herrick, 1994. A family of transposase genes in transposons found in prokaryotes, multicellular eukaryotes and ciliated protozoans. Proc. Natl. Acad. Sci., USA 91: 942–946.
Entwistle, P. F., J. S. Cory, M. J. Bailey & S. Higgs, 1993.Bacillus thuringiensis, an environmental biopesticide: theory and practice. J. Wiley & Sons, Chichester, England.
Fayet, O., P. Ramond, P. Polard, M.-F. Prère & M. Chandler, 1990. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol. Microbiol. 4: 1771–1777.
Gamel, P. H. & J.-C. Piot, 1992. Characterization and properties of a novel plasmid vector forBacillus thuringiensis displaying compatibility with host plasmids. Gene 120: 17–26.
Gelernter, W. & G. E. Schwab, 1993. Transgenic bacteria, viruses, algae and other microorganisms asBacillus thuringiensis toxin delivery systems, pp. 89–104 inBacillus thuringiensis, an environmental biopesticide: theory and practice, edited by P. F. Entwistle, J. S. Cory, M. J. Bailey and S. Higgs. J. Wiley & Sons, Chichester, England.
González, J. M., Jr., B. J. Brown & B. C. Carlton, 1982. Transfer ofBacillus thuringiensis plasmids coding for δ-endotoxin among strains ofB. thuringiensis andB. cereus. Proc. Natl. Acad. Sci. USA 79: 6951–6955.
Green, B. D., L. Battisti & C. B. Thorne, 1989. Involvement of Tn4430 in transfer ofBacillus anthracis plasmids mediated byBacillus thuringiensis plasmid pXO12. J. Bacteriol. 171: 104–113.
Hallet, B., R. Rezsöhazy & J. Delcour, 1991. IS231 A fromBacillus thuringiensis is functional inEscherichia coli: transposition and insertion specificity. J. Bacteriol. 173: 4526–4529.
Hallet, B., R. Rezsöhazy, J. Mahillon & J. Delcour, 1994. IS231 A insertion specificity: consensus sequence and DNA bending at the target site. Mol. Microbiol. 13 (in press).
Hodgman, T. C., Y. Ziniu, J. Shen & D. J. Ellar, 1993. Identification of a cryptic gene associated with an insertion sequence not previously identified inBacillus thuringiensis. FEMS Microbiol. Lett. 114: 23–30.
Höfte, H. & H. R. Whiteley, 1989. Insecticidal crystal proteins ofBacillus thuringiensis. Microbiol. Rev. 53: 242–255.
Ji, H., D. P. Moore, M. A. Blomberg, L. T. Braiterman, D. F. Voytas, G. Natsoulis & J. D. Boeke, 1993. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73: 1007–1018.
Khan, E., J. P. G. Mack, R. A. Katz, J. Kulkosky & A. M. Skalka, 1991. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 19: 851–860.
Klaer, R., S. Kühn, E. Tillman, H.-J. Fritz & P. Starlinger, 1981. The sequence of IS4. Mol. Gen. Genet. 181: 169–175.
Kleckner, N., 1989. Transposon Tn10, pp. 227–268 in Mobile DNA, edited by D. E. Berg and M. M. Howe. American Society for Microbiology, Washington, D.C.
Kothary, R. K., D. Jones & E. P. M. Candido, 1985. IS186: anEscherichia coli insertion element isolated from a cDNA library. J. Bacteriol. 164: 957–959.
Kronstad, J. W. & H. R. Whiteley, 1984. Inverted repeat sequences flank aBacillus thuringiensis crystal protein gene. J. Bacteriol. 160: 95–102.
Kronsad, J. W. & H. R. Whiteley, 1986. Three classes of homologousBacillus thuringiensis crystal-protein genes. Gene 43: 29–40.
Kulkosky, J., K. S. Jones, R. A. Katz, J. P. G. Mack & A. M. Skalka, 1992. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial Insertion Sequence transposases. Mol. Cell. Biol. 12: 2331–2338.
Lereclus, D., A. Delécluse & M.-M. Lecadet, 1993. Diversity ofBacillus thuringiensis toxins and genes, pp. 37–69 inBacillus thuringiensis, an environmental biopesticide: theory and practice, edited by P. F. Entwistle, J. S. Cory, M. J. Bailey and S. Higgs. J. Wiley & Sons, Chichester, England.
Lereclus, D., S. Guo, V. Sanchis & M.-M. Lecadet, 1988. Characterization of twoBacillus thuringiensis plasmids whose replication is thermosensitive inB. subtilis. FEMS Microbiol. Lett. 49: 417–422.
Lereclus, D., J. Mahillon, G. Menou & M.-M. Lecadet, 1986. Identification of Tn4430, a transposon ofBacillus thuringiensis functional inEscherichia coli. Mol. Gen. Genet. 204: 52–57.
Lereclus, D., G. Menou & M.-M. Lecadet, 1983. Isolation of a DNA sequence related to several plasmids fromBacillus thuringiensis after a mating involving theStreptococcus faecalis pAMß1. Mol. Gen. Genet. 191: 307–313.
Lereclus, D., J. Ribier, A. Klier, G. Menou & M.-M. Lecadet, 1984. A transposon-like structure related to the δ-endotoxin gene ofBacillus thuringiensis. EMBO J. 3: 2561–2567.
Lereclus, D., M. Vallade, J. Chaufaux, O. Arantes & S. Rambaud, 1992. Expansion of insecticidal host range ofBacillus thuringiensis by in vivo genetic recombination. Biotechnology 10: 418–421.
Mahillon, J., F. Hespel, A.-M. Pierssens & J. Delcour, 1988. Cloning and partial characterization of three small cryptic plasmids fromBacillus thuringiensis. Plasmid 19: 169–173.
Mahillon, J. & D. Lereclus, 1988. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 7: 1515–1526.
Mahillon, J. & J. Seurinck, 1988. Complete nucleotide sequence of pGI2, aBacillus thuringiensis plasmid containing Tn4430. Nucleic Acids Res. 16: 11827–11828.
Mahillon, J., J. Seurinck, J. Delcour & M. Zabeau, 1987. Cloning and nucleotide sequence of different iso-IS231 elements and their structural association with the Tn4430 transposon inBacillus thuringiensis. Gene 51: 187–196.
Mahillon, J., J. Seurinck, L. Van Rompuy, J. Delcour & M. Zabeau, 1985. Nucleotide sequence and structural organization of an insertion sequence element (SI231) fromBacillus thuringiensis strain berliner 1715. EMBO J. 4: 3895–3899.
Menou, G., J. Mahillon, M.-M. Lecadet & D. Lereclus, 1990. Structural and genetic organization of IS232, a new insertion sequence ofBacillus thuringiensis. J. Bacteriol. 172: 6689–6696.
Mizuuchi, K., 1992. Polynucleotidyl transfer reactions in transpositional DNA recombination. J. Biol. Chem. 267: 21273–21276.
Mollet, B., S. Iida, J. Sepherd & W. Arber, 1983. Nucleotide sequence of IS26, a new prokaryotic mobile genetic element. Nucleic Acids Res. 11: 6319–6330.
Polard, P., M.-F. Prère, M. Chandler & O. Fayet, 1991. Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J. Mol. Biol. 222: 465–477.
Rezsöhazy, R., B. Hallet & J. Delcour, 1992. IS231 D, E and F, three new insertion sequences inBacillus thuringiensis: extension of the IS231 family. Mol. Microbiol. 6: 1959–1967.
Rezsöhazy, R., B. Hallet, J. Delcour & J. Mahillon, 1993a. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol. Microbiol. 9: 1283–1295.
Rezöhazy, R., B. Hallet, J. Mahillon & J. Delcour, 1993b. IS231 V and W fromBacillus thuringiensis subsp.israelensis, two distant members of the IS231 family of insertion sequences. Plasmid 30: 141–149.
Ryan, M., J. D. Johnson & L. A. Bulla, Jr., 1993. Insertion sequence elements inBacillus thuringiensis subsp.darmstadiensis. Can. J. Microbiol. 39: 649–658.
Sanchis, V., D. Lereclus, G. Menou, J. Chaufaux & M.-M. Lecadet, 1988. Multiplicity of δ-endotoxin genes with different insecticidal specificities inBacillus thurnngiensis aizawai 7.29. Mol. Microbiol. 2: 393–404.
Schnepf, H. E. & H. R. Whiteley, 1981. Cloning and expression of theBacillus thuringiensis crystal protein gene inEscherichia coli. Proc. Natl. Acad. Sci. USA 78: 2893–2897.
Sekine, Y. & E. Ohtsubo, 1989. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc. Natl. Acad. Sci. USA 86: 4609–4613.
Smith, G. P., D. J. Ellar, S. J. Keeler & C. E. Seip, 1994. Nucleotide sequence and analysis of an Insertion Sequence fromBacillus thuringiensis related to IS150. Plasmid (in press).
Trieu-Cuot, P. & P. Courvalin, 1984. Nucleotide sequence of the transposable element IS15. Gene 30: 113–120.
van Luenen, H. G. A. M. & R. H. A. Plasterk, 1994. Target site choice of the related transposable elements Tc1 and Tc3 ofCaenorhabditis elegans. Nucleic Acids Res. 22: 262–269.
Vögele, K., E. Schwartz, C. Welz, E. Schiltz & B. Rak, 1991. Highlevel ribosomal frameshifting directs the synthesis of IS150 gene products. Nucleic Acids Res. 19: 4377–4385.
Weiss, R. B., D. M. Dunn, A. E. Dahlberg, J. F. Atkins & R. F. Gesteland, 1988. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis inEscherichia coli. EMBO J. 7: 1503–1507.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Mahillon, J., Rezsöhazy, R., Hallet, B. et al. IS231 and otherBacillus thuringiensis transposable elements: A review. Genetica 93, 13–26 (1994). https://doi.org/10.1007/BF01435236
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01435236