Summary
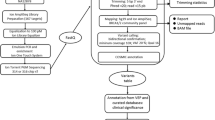
Chromosome 17q12-21 is known to contain a gene (or genes) which confers susceptibility to early-onset breast cancer and ovarian cancer (BRCA1). Identification and isolation of BRCA1 will likely provide the basis for increased understanding of the pathogenesis of breast and ovarian cancer, the development of targeted diagnostic and therapeutic approaches, and a means of screening women at risk of being BRCA1 mutation carriers. Genetic and physical maps of the BRCA1 candidate region have been largely completed and efforts are being directed at identification of candidate genes from within this region. We have begun the task of identifying transcripts from this region employing three complementary strategies. These include: 1) direct cDNA screening with cosmids derived from the BRCA1 region; 2) exon amplification; and 3) magnetic bead capture. Transcripts identified using these approaches are being characterized for: 1) tissue expression pattern; 2) the presence of genomic rearrangement in DNA derived from affected members of families believed to show linkage between breast cancer and genetic markers in the BRCA1 candidate interval; 3) altered size and/or expression pattern in RNA prepared from such individuals; and 4) homology to known genes or functional motifs. Germline mutations in affected individuals from these families will serve as presumptive evidence of BRCA1 identity.
Similar content being viewed by others
References
Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King M-C: Linkage of early onset breast cancer to chromosome 17q21. Science 250:1684–1689, 1990.
Narod SA, Feuteun J, Lynch HT, Watson P, Conway T, Lynch J, Lenoir GM: Familial breast-ovarian cancer locus on chromosome 17q12-23. Lancet 338: 82–83, 1991.
Easton DF, Bishop DT, Ford D, Crockford GP, and the Breast Cancer Linkage Consortium: Genetic linkage analysis in familial breast and ovarian cancer — results from 214 families. Am J Hum Genet 52:678–701, 1993.
Claus EB, Risch N, Thompson WD: Genetic analysis of breast cancer in the cancer and steroid hormone study. Am J Hum Genet 48:232–242, 1991.
Colditz GA, Willett WC, Hunter DJ, Stampfer MJ, Manson JE, Hennekens CH, Rosner BA: Family history, age and risk of breast cancer. JAMA 270: 338–343, 1993.
Slattery ML, Kerber RA: A comprehensive evaluation of family history and breast cancer risk: the Utah Population Database. JAMA 270:1563–1568, 1993.
Weber BL, Garber JE: Family history and breast cancer: probabilities and possibilities. JAMA 270: 1602–1603, 1993.
Knudson AG: Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA 68:820–823, 1971.
Smith SA, Easton DF, Evans DGR, Ponder BAJ: Allele losses in the region 17q12-21 in familial breast and ovarian cancer involve the wild-type chromosome. Nature Genetics 2:128–131, 1991.
Chamberlain JS, Boehnke M, Frank TS, Kiousis S, Xu J, Guo S-W, Hauser ER, Norum RA, Helmbold EA, Markel DS, Keshavarzi SM, Jackson CE, Calzone K, Garber J, Collins FS, Weber BL: BRCA1 maps proximal to D17S579 on chromosome 17q21 by genetic analysis. Am J Hum Genet 52:792–798, 1993.
Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW: Identification of a chromosome 18q gene that is altered in colorectal cancers. Science 247:49–56, 1990.
Lee WH, Bookstein R, Hong F, Young LJ, Shew JY, Lee EY: Human retinoblastoma susceptibility gene: cloning, identification and sequence. Science 235: 1394–1399, 1987.
Collins FS: Positional cloning: Let's not call it reverse any more. Nature Genetics 1:3–6, 1992.
Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N: Identification of the cystic fibrosis gene: Chromosome walking and jumping. Science 245: 1059–1065, 1989.
Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, Fountain JW, Brereton A, Nicholson J, Mitchell AL: Type I neurofibromatosis gene: Identification of a large transcript disrupted in three NFI patients. Science 249:181–186, 1990.
The Huntington's Disease Collaborative Research Group: A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's Disease chromosome. Cell 72:971–983, 1993.
Kinzler KW, Nilbert MC, Su L-K, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hedge P, McKechnie D: Identification of FAP locus genes from chromosome 5q21. Science 253:661–665, 1991.
Abel KJ, Boehnke M, Prahalad M, Ho P, Flejter WL, Watkins M, VanderStoep J, Chandrasekharappa SC, Collins FS, Glover TW, Weber BL: A radiation hybrid map of the BRCA1 region of chromosome 17q12-21. Genomics 17:632–641, 1993.
Flejter WL, Barcroft CL, Guo S-W, Lynch ED, Boehnke M, Chandrasekharappa SC, Hayes S, Collins FS, Weber BL, Glover TW: Multi-color FISH mapping with Alu-PCR amplified YAC clone DNA determines the order of markers in the BRCA1 region on chromosome 17q12-12. Genomics 17:624–631, 1993.
Anderson LA, Friedman L, Osborne-Lawrence S, Lynch E, Weissenbach J, Bowcock A, King M-C: Highdensity genetic map of the BRCA1 region of chromosome 17q12-21. Genomics 17:618–623, 1993.
Bowcock AM, Anderson LA, Black DM, Lawrence SO, Hall JM, King M-C: Polymorphisms in THRA1 and D17S183 flank less than 4 cM interval for the breast-ovarian cancer gene on chromosome 17q. Am J Hum Genet (in Press).
Simard J, Feuteun J, Lenoir G, Tonin P, Normand T, Luu The V, Vivier A, Lasko D, Morgan K, Rouleau GA, Lynch H, Labrie F, Narod SA: Genetic mapping of the breast-ovarian cancer syndrome to a small interval on chromosome 17q12-21: exclusion of candidate genes EDH17B2 and RARA. Human Molecular Genetics 2:1193–1199, 1993.
Buckler AJ, Chang DD, Graw SL, Brook JD, Haber DA, Sharp PA, Housman DE: Exon amplification: A strategy to isolate mammalian genes based on RNA splicing. Proc Natl Acad Sci USA 88:4005–4009, 1991.
Lovett M, Kere J, Hinton LM: Direct selection: A method for the isolation of cDNAs encoded by large genomic regions. Proc Natl Acad Sci USA 88:9628–9632, 1991.
Tagle DA, Swaroop M, Lovett M, Collins FS: Magnetic bead capture of expressed sequences within large genomic segments. Nature 161:751–756, 1993.
Buckler AJ, Statler CJ, Rutter JL, Murrell J, Trofatter JA, McCormick MK, Deaven L, Mayzis RK, Gusella JF: Chromosome specific libraries of exons and high resolution transcription maps [abstract]. Am J Hum Genet 53:A192.
North MA, Sanseau P, Bucker AJ, Church D, Jackson A, Patel K, Trowsdale J, Lehrach H: Efficiency and specificity of gene isolation by exon amplification. Mammalian Genome 4:466–474, 1993.
Krizman DB, Berget SM: 3'-terminal exon trapping: Identification of genes from vertebrate DNA. Focus 15:106–108, 1993.
Cotton RG, Rodrigues NR, Campbell RD: Reactivity of cytosine and thymine in single base pair mismatches with hydroxylamine and osmium tetraoxide and its application to the study of mutations. Proc Natl Acad Sci USA 85:4397–4401, 1988.
Orita M, Suziki Y, Sekiya T, Hayashi K: Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics 5:874–879, 1989.
Kinzler KW, Nilbert MC, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hamilton SR, Hedge P, Markham A, Carlson M, Joslyn G, Groden J, White R, Miki Y, Miyoshi Y, Nishisho I, Nakamura Y: Identification of a gene located at chromosome 5q21 that is mutated in colorectal cancers. Science 251:1366–1370, 1991.
Sato T, Saito H, Swensen J, Olifant A, Wood C, Danner D, Sakamoto T, Takita K, Kasumi F, Miki Y, Skolnik M, Nakamura Y: The human prohibitin gene located on chromosome 17q21 is mutated in sporadic breast cancer. Cancer Res 52:1643–1646, 1992.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Weber, B.L., Abel, K.J., Couch, F.J. et al. Transcript identification in the BRCA1 candidate region. Breast Cancer Res Tr 33, 115–124 (1995). https://doi.org/10.1007/BF00682719
Issue Date:
DOI: https://doi.org/10.1007/BF00682719