Summary
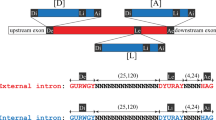
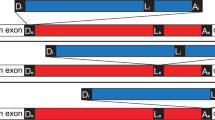
We have shown that the second intron of thePodospora mitochondrial gene coding for cytochromeb (Cytb 12) splices autocatalytically, using in vitro transcripts generated from the T7 promoter. The reaction takes place at 37° C in the presence of 50 mM TRIS-HCl pH 7.5, 60 mM MgC12 and 1 mM GTP but shows a low efficiency even at high KCl concentrations of up to 1.2 M. Under these conditions, intron bI2 follows the conventional pathway of group I splicing, and all characteristic products, with regard to both transesterification and hydrolysis, could be identified. Moreover, the intron is capable of undergoing cyclization, thereby releasing the noncoded G and one additional nucleotide (U) from the 5′ end. The 5′ cleavage site is preceded by the same two nucleotides, indicating a base-pairing at the same site of the internal guide sequence (IGS) for both splicing and cyclization (“one-binding-site model”). In addition, products resulting from site-specific hydrolysis 138 nucleotides downstream of the 5′ splice site were detected. Unusually, the shortened intron is also able to form a circular RNA and an alternative sequence that aligns the cyclization site to the catalytic core of the intron must be assumed.
Similar content being viewed by others
References
Burke JM, Belfort M, Cech TR, Davies RW, Schweyen RJ, Shub DA, Szostak JW, Tabak HF (1987) Structural conventions for group I introns. Nucleic Acids Res 15:7217–7221
Casabadan MJ, Chou J, Cohen SN (1980) In vitro gene fusion that join an enzymatically active beta-galactosidase segment to aminoterminal fragments of exogenous proteins —Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol 143: 971–980
Cech TR (1987) The chemistry of self-splicing RNA and RNA enzymes. Science 236:1532–1539
Cech TR (1988) Conserved sequences and structures of group I introns: building an active site for RNA catalysis — a review. Gene 73:259–271
Cech TR (1990) Self-splicing of group I introns. Annu Rev Biochem 59:543–568
Cech TR, Zaug AJ, Grabowski PJ (1981) In vitro splicing of the ribosomal RNA precursor ofTetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 27:487–496
Cummings DJ, Michel F, McNally KL (1989) DNA sequence analysis of the apocytochrome b gene ofPodospora anserina: a new family of intronic open reading frame. Curr Genet 16:407–418
Cummings DJ, McNally KL, Domenico JM, Matsuura ET (1990) The complete DNA sequence of the mitochondrial genome ofPodospora anserina. Curr Genet 17:375–402
Davies RW, Waring RB, Ray JA, Brown TA, Scazzocchio C (1982) Making ends meet: a model for RNA splicing in fungal mitochondria. Nature 300:719–724
Ehrenman K, Pedersen-Lane J, West D, Herman R, Maley F, Belfort M (1986) Processing of phage T4 td-encoded RNA is analogous to the eukaryotic group I splicing pathway. Proc Natl Acad Sci USA 83:5875–5879
Garriga G, Lambowitz AM (1984) RNA splicing inNeurospora mitochondria: self-splicing of a mitochondrial intron in vitro. Cell 39:631–641
van der Horst G, Tabak HF (1985) Self-splicing of yeast mitochondrial ribosomal and messenger RNA precursors. Cell 40:759–766
Inoue T, Sullivan FX, Cech TR (1986) New reactions of the ribosomal RNA precursor ofTetrahymena and the mechanism of self-splicing. J Mol Biol 189:143–165
Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence ofTetrahymena. Cell 31:147–157
Kück U, Stahl U, Esser K (1981) Plasmid-like DNA is a part of mitochondrial DNA inPodospora anserina. Curr Genet 3:151–156
Michel F, Dujon B (1983) Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J 2:33–38
Michel F, Westhof E (1990) Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol 216:585–610
Michel F, Jacquier A, Dujon B (1982) Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie 64:867–881
Partono S, Lewin AS (1988) Autocatalytic activities of intron 5 of the cob gene of yeast mitochondria. Mol Cell Biol 8:2562–2571
Partono S, Lewin AS (1990) Splicing of COB intron 5 requires pairing between the internal guide sequence and both flanking exons. Proc Natl Acad Sci USA 87:8192–8196
Partono S, Lewin AS (1991) The rate and specificity of a group I ribozyme are inversely affected by choice of the monovalent salt. Nucleic Acids Res 19:605–609
Schmidt U, Riederer B, Mörl M, Schmelzer C, Stahl U (1990) Self-splicing of the mobile group II intron of the filamentous fungusPodospora anserina (COI I1) in vitro. EMBO Jg:2289–2298
Sullivan FX, Cech TR (1985) Reversibility of cyclization of theTetrahymena rRNA intervening sequence: implication for the mechanism of splice site choice. Cell 42:639–648
Tabak HF, van der Horst G, Osinga KA, Arnberg AC (1984) Splicing of large ribosomal precursor RNA and processing of intron RNA in yeast mitochondria. Cell 39:623–629
Tabak HF, van der Horst G, Kamps AMJE, Arnberg AC (1987) Interlocked RNA circle formation by a self-splicing yeast mitochondrial group I intron. Cell 48:101–110
Waring RB, Davies RW (1984) Assessment of a model for intron secondary structure relevant to RNA self-splicing — a review. Gene 28:277–291
Zaug AJ, Cech TR (1982) The IVS excised from the rRNA precursor ofTetrahymena contains a 5′-terminal guanosine residue not encoded by the DNA. Nucleic Acids Res 10:2823–2838
Zaug AJ, Cech TR (1985) Oligomerization of intervening sequence RNA molecules in the absence of proteins. Science 229:1060–1064
Zaug AJ, Grabowski PJ, Cech TR (1983) Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature 301:578–583
Zaug AJ, Kent JR, Cech TR (1984) A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science 224:574–578
Zaug AJ, Kent JR, Cech TR (1985) Reactions of the intervening sequence of theTetrahymena ribosomal ribonucleic acid precursor: pH dependence of cyclization and site-specific hydrolysis. Biochemistry 24:6211–6218
Author information
Authors and Affiliations
Additional information
Communicated by C. van den Hondel
Rights and permissions
About this article
Cite this article
Schmidt, U., Budde, E. & Stahl, U. Self-splicing of a mitochondrial group I intron from the cytochromeb gene of the ascomycetePodospora anserina . Molec. Gen. Genet. 233, 71–80 (1992). https://doi.org/10.1007/BF00587563
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00587563