Summary
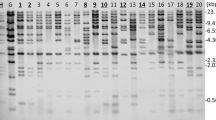
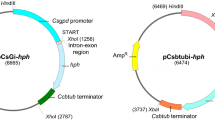
Conditions are described for the efficient isolation and regeneration of protoplasts of a fungal pathogen of brassicas, Leptosphaeria maculans. Treatment of the protoplasts with DNA of the plasmid pAN7-1 (containing an E. coli hygromycin phosphotransferase gene with Aspergillus nidulans expression signals) and plating under selective conditions resulted in the formation of hygromycin 13-resistant colonies. Southern blot analysis of resistant colonies indicated that single copies of the plasmid had integrated into different sites in the genome. In twelve of the transformants analysed so far, the integration is stable through mitosis. The demonstration of efficient transformation is an essential first step in the molecular analysis of pathogenicity of this commercially important pathogen.
Similar content being viewed by others
References
Gabrielson RL (1983) Seed Sci Technol 11:749–780
Hammond KM, Lewis BG, Musa TM (1985) Plant Pathol 34:557–565
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York (A laboratory manual)
McGee DC, Petrie GA (1987) Phytopathology 68:625–630
Mishra NC (1985) Adv Genet 23:73–178
Mithen RF, Lewis BG, Fenwick GR (1986) Trans Brit Mycol Soc 87(3):433–440
Musa TM (1981) Host-pathogen interaction between Brassica napus L. spp oleifera (Metzger) Sink, and Leptosphaeria maculans (Desm.) Cos. et de Not. PhD Thesis, University of East Anglia, Norwich, UK
Oliver RP, Roberts IN, Harling R, Kenyon L, Punt PJ, Dingemanse MA, van den Hondel CAMJJ (1987) Curr Genet 12:231–233
Peberdy JF (1979) Annu Rev Microbiol 33:21–39
Peberdy JF (1987) Microbiol Sci 4:108–114
Pittinger RC, Wolfe RN, Hoehn MM, Marks PN, Daily WA, McGuire JM (1953) Antibiot Chemother 3:1268–1282
Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CAMJJ (1987) Gene 56:117–124
Queener SW, Ignolia TD, Skatrud PL, Chapman JL, Kaster KR (1985) In: Schlessinger D (ed) Microbiology. Am Soc Microbiol 468–471
Raeder U, Broda P (1985) Lett Appl Microbiol 1:17–20
Turgeon BG, Garber RC, Yoder OC (1985) Mol Gen Genet 201:450–453
Turgeon BG, Garber RC, Yoder OC (1986) Abstract from the 3rd international symposium for molecular genetics of plantmicrobe interactions. McGill University, Montreal, p 67
Vollmer SJ, Yanofsky C (1986) Proc Natl Acad Sci USA 83:4869–4873
Yelton MM, Hamer JE, Timberlake WE (1984) Proc Natl Acad Sci USA 81:1470–1474
Yoder OC, Weltring K Turgeon BG, VanElten HD (1985) Biology and molecular biology of plant-pathogen interactions. Proc Nato ARW, Dillington College, p 15
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Farman, M.L., Oliver, R.P. The transformation of protoplasts of Leptosphaeria maculans to hygromycin B resistance. Curr Genet 13, 327–330 (1988). https://doi.org/10.1007/BF00424427
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00424427