Summary
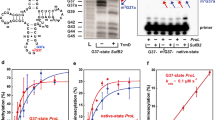
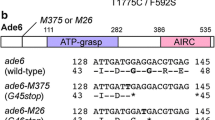
The yeast invertase structural gene SUC2 has two naturally occurring alleles, the active one and a silent allele called suc2°. Strains carrying suc2° are unable to ferment sucrose and do not show detectable invertase activity. We have isolated suc2° and found an amber codon at position 232 of 532 amino acids. However, transformants carrying suc2° on a multicopy plasmid were able to ferment sucrose and showed detectable invertase activity. Full-length invertase was found in gels stained for active invertase and in immunoblots. Therefore we concluded that the amber codon is occasionally read as an amino acid. The calculated frequency of read-through is about 4% of all translation events.
Similar content being viewed by others
References
Aviv H, Leder P (1972) Purification of biologically active globin mRNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci USA 69:1408–1412
Baserga SJ, Benz EJ (1988) Nonsense mutations in the human β-globin gene affect mRNA metabolism. Proc Natl Acad Sci USA 85:2056–2060
Beggs JD (1978) Transformation of yeast by a replicating plasmid. Nature 275:447–452
Beggs JD (1981) Multiple-copy yeast plasmid vectors. Alfred Benzon Symp 16:383–390
Bossi L, Roth JR (1980) The influence of codon context on genetic code translation. Nature 286:123–127
Burnette WN (1981) “Western blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem 112:195–203
Carlson M, Botstein D (1982) Two differentially regulated mRNAs with different 5′-ends encode secreted and intracellular forms of yeast invertase. Cell 28:145–154
Carlson M, Botstein D (1983) Organization of the SUC-gene family in Saccharomyces. Mol Cell Biol 3:351–359
Carlson M, Osmond BC, Botstein D (1981) Genetic evidence for a silent SUC-gene in yeast. Genetics 98:41–54
Carlson M, Celenza J, Eng FJ (1985) Evolution of the dispersed SUC-gene family of Saccharomyces by rearrangement of chromosome telomeres. Mol Cell Biol 5:2894–2902
Castillo Agudo L del, Zimmermann FK (1986) A silent gene allelic to yeast invertase structural gene SUC2 can be activated by mutation to direct the formation of a new type of invertase. Curr Microbiol 14:165–167
Denis CL, Ferguson J, Young ET (1981) A positive regulatory gene is required for accumulation of functional mRNA for the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. J Mol Biol 148:355–368
Feinstein SI, Altman S (1978) Context effects of nonsense codon suppression in Escherichia coli. Genetics 88:201–219
Gallwitz D, Sures J (1980) Structure of a split gene: Complete nucleotide sequence of the actin gene in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 77:2546–2550
Gascón S, Ottolenghi P (1972) Influence of glucose concentration of the medium on the invertase content of a strain of Saccharomyces bearing the SUC2 gene. C R Trav Lab Carlsberg 39:15–24
Goldstein A, Lampen JO (1975) β-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol 42:504–511
Grossmann MK, Zimmermann FK (1979) The structural genes of internal invertases in Saccharomyces cerevisiae. Mol Gen Genet 175:223–229
Hohmann S (1987) Physiologische und molekulargenetische Untersuchungen zur Saccarosevergärung der Hefe Saccharomyces cerevisiae PhD thesis, Technische Hochschule Darmstadt
Hohmann S, Zimmermann FK (1986) Cloning and expression on a multicopy vector of five invertase genes of Saccharomyces cerevisiae. Curr Genet 11:217–225
Kohli J, Grosjean H (1981) Usage of the three termination codons: Complication and analysis of the known eucaryotic and procaryotic translation termination sequences. Mol Gen Genet 182:430–439
Kunes S, Ma H, Overbye K, Fox MS, Botstein D (1987) Fine structure recombinational analysis of cloned genes using yeast transformation. Genetics 115:73–81
Losson L, Lacroute F (1979) Interference of nonsense mutations with eucaryotic messenger RNA stability. Proc Natl Acad Sci USA 76:5134–5137
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Murray AW, Szostak JW (1983) Pedigree analysis of plasmid segregation in yeast. Cell 34:961–970
Pelsy F, Lacroute F (1984) Effect of ochre nonsense mutations on yeast URA1 mRNA stability. Curr Genet 8:277–282
Ryoji M, Hsia K, Kaji A (1983) Read-through translation. Trends Biochem Sci 8:88–90
Struhl K, Stinchcomb DT, Scherer S, Davis RW (1979) High frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA 76:1035–1039
Taussig R, Carlson M (1983) Nucleotide sequence of the yeast SUC2 gene for invertase. Nucleic Acids Res 11:1943–1954
Tautz D, Renz M (1983) An optimized freeze squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem 48:661–671
Weiss WA, Friedberg EC (1986) Normal yeast tRNA glncag can suppress amber codons and is encoded by an essential gene. J Mol Biol 192:725–735
Winston F, Chumley F, Fink GR (1983) Eviction and transplacement of mutant genes in yeast. Methods Enzymol 101:211–228
Zamenhoff S (1957) Preparation and assay of deoxyribonucleic acids from animal tissues. Methods Enzymol 3:696–704
Author information
Authors and Affiliations
Additional information
Communicated by C.P. Hollenberg
Rights and permissions
About this article
Cite this article
Gozalbo, D., Hohmann, S. The naturally occurring silent invertase structural gene suc2° contains an amber stop codon that is occasionally read through. Mol Gen Genet 216, 511–516 (1989). https://doi.org/10.1007/BF00334398
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00334398