Summary
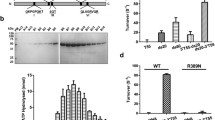
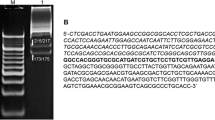
The activity of Ustilago maydis DNAse I, an enzyme implicated in genetic recombination, on DNA substrates containing unpaired or mismatched bases, was examined. The enzyme nicked supercoiled PM-2 molecules, converting these to relaxed circular and linear molecules. Discrete double stranded linear fragments smaller than unit length were also observed after digestion at high enzyme concentration. Heteroduplex molecules were constructed using ϕ80 bacteriophage derivatives which contained single base substitutions within the E. coli tRNA tyr1 gene. Single and double stranded nicking at or near the single mismatched site was observed with three out of the five pairs of heteroduplexes.
Similar content being viewed by others
References
Ahmad, A., Holloman, W.K., Holliday, R.: Nuclease that preferentially inactivates DNA containing mismatched bases. Nature (Lond.) 258, 54–56 (1975)
Badman, R.: DNase-deficient mutants of Ustilago maydis with altered recombination frequences. Genet. Res. 20, 213–229 (1972)
Beard, P., Morrow, J., Berg, P.: Cleavage of circular, superhelical simian virus 40 DNA to a linear duplex by S1 nuclease. J. Virol. 12, 1303–1313 (1973)
Brack, C., Bickle, T.A., Yuan, R.: The relation of single-stranded regions in bacteriophage PM-2 supercoiled DNA to the early melting sequences. J. molec. Biol. 96, 693–702 (1975)
Fogel, S., Mortimer, R. K.: Informational transfer in meiotic gene conversion. Proc. nat. Acad. Sci. (Wash.) 62, 96–103 (1969)
Germond, J.E., Vogt, V.M., Hirt, B.: Characterization of the single-strand-specific nuclease S1 activity on double-stranded supercoiled polyoma DNA. Europ. J. Biochem. 43, 591–600 (1974)
Godson, G.N.: Action of the single-stranded DNA specific nuclease S1 on double-stranded DNA. Biochim. biophys. Acta (Amst.) 308, 59–67 (1973)
Goodman, H.M., Olson, M.V., Hall, B.D.: Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc. nat. Acad. Sci. (Wash.) 74, 5453–5457 (1977)
Gray, H.B., Jr., Upholt, W.B., Vinograd, J.: A buoyant method for the determination of the superhelix density of closed circular DNA. J. molec. Biol. 62, 1–19 (1971)
Greenfield, L., Simpson, L., Kaplan, D.: Conversion of closed circular DNA molecules to single-nicked molecules by digestion with DNase I in the presence of ethidium bromide. Biochim. biophys. Acta (Amst.) 407, 365–375 (1975)
Griffin, B.E., Fried, M., Cowie, A.: Polyoma DNA: A physical map. Proc. nat. Acad. Sci. (Wash.) 71, 2077–2081 (1974)
Holliday, R.: a mechanism for gene conversion in fungi. Genet. Res. 5, 282–304 (1964)
Holliday, R.: Studies on mitotic gene conversion in Ustilago maydis. Genet. Res. 8, 323–337 (1966)
Holliday, R., Dickson, J.M.: The detection of post-meiotic segregation without tetrad analysis in Ustilago maydis. Evidence that a mutation defective in excision of pyrimidine dimers can repair mismatched bases in hybrid DNA. Molec gen. Genet. 153, 331–336 (1977)
Holliday, R., Holloman, W.K., Banks, G.R., Unrau, P., Pugh, J.E.: Genetic and biochemical studies of recombination in Ustilago maydis. In: Mechanisms in recombination (R.F. Grell, ed.) pp. 239–262 New York: Plenum Press 1974
Holloman, W.K.: Studies on a nuclease from Ustilago maydis. II. Substrate specificity and mode of action of the enzyme. J. biol. Chem. 248, 8114–8119 (1973)
Holloman, W.K., Holliday, R.: Studies on a nuclease from Ustilago maydis. I. Purification, properties, and implication in recombination of the enzyme. J. biol. Chem. 248, 8107–8113 (1973)
Kato, A.C., Bartok, K., Fraser, M.J., Denhardt, D.T.: Sensitivity of superhelical DNA to a single-strand specific endonuclease. Biochim. biophys. Acta (Amst.) 308, 68–78 (1973)
Kelly, T.J., Smith, H.O.: A restriction enzyme from Hemophilus influenzae II Base sequence of the recognition site. J. molec. Biol. 51, 393–409 (1970)
Landy, A., Foeller, C., Ross, W.: DNA fragments carrying genes for tRNA tyr1 . Nature (Lond.) 249, 738–742 (1974)
Leblon, G.: Mechanism of gene conversion in Ascobolus immersus. I. Existence of a correlation between the origin of mutants induced by different mutagens and their conversion spectrum. Molec. gen. Genet. 115, 36–48 (1972a)
Leblon, G.: Mechanism of gene conversion in Ascobolus immersus. II. The relationships between the genetic alterations in b1 or b2 mutants and their conversion spectrum. Molec. gen. Genet. 116, 322–335 (1972b)
Leblon, G., Rossignol, J-L.: Mechanism of gene conversion in Ascobolus immersus. III. The interaction of heteroalleles in the conversion process. Molec. gen. Genet. 122, 165–182 (1973)
Lindegren, C.C.: Gene conversion in Saccharomyces. J. Genet. 51, 625–637 (1953)
Mechali, M., de Recondo, A-M., Girard, M.: Action of the S1 endonuclease from Aspergillus oryzae on simian virus 40 supercoiled component I DNA. Biochem. biophys. Res. Commun. 54, 1306–1320 (1973)
Meselson, M.S.: The duplication and recombination of genes. In: Ideas in modern biology (J.A. Moore, ed.) pp. 5–16. New York: The Natural History Press, Garden City 1965
Mitchell, M.B.: Aberrant recombination of pyridoxine mutants of Neurospora. Proc. nat. Acad. Sci. (Wash.) 41, 215–220 (1955)
Murray, K., Murray, N.E.: Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease III of Haemophilus influenzae and restriction endonuclease I of Escherichia coli. J. molec. Biol. 98, 551–564 (1975)
Panet, A., Van de Sande, J.H., Loewen, P.C., Khorana, H.G., Raae, A.J., Lillehaug, J.R., Kleppe, K.: Physical characterization and simultaneous purification of bacteriophage T4 induced polynucleotide kinase, polynucleotide ligase, and deoxyribonucleic acid polymerase. Biochem. 12, 5045–5050 (1973)
Philippsen, P., Streeck, R.E., Zachau, H.G.: Defined fragments of calf, human, and rat DNA produced by restriction nucleases. Europ. J. Biochem. 45, 479–488 (1974)
Randerath, K., Randerath, E.: Thin layer separation methods for nucleic acid derivatives. In: Methods in enzymology, XII A. (L. Grossman and K. Moldave, ed.), pp. 323–347. New York: Academic Press 1967
Richardson, J.P.: Attachment of nascent RNA molecules to superhelical DNA. J. molec. Biol. 98, 565–579 (1975)
Rossignol, J-L.: Existence of homogeneous categories of mutants exhibiting various conversion patterns in gene 75 of Ascobolus immersus. Genetics 63, 795–805 (1969)
Sharp, P.A., Sudgen, B., Sambrook, J.: Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose-ethidium bromide electrophoresis. Biochem. 12, 3055–3063 (1975)
Shenk, T.E., Rhodes, C., Rigby, P.W.J., Berg, P.: Biochemical method for mapping mutational alteration in DNA with S1 nuclease: The location of deletions and temperature-sensitive mutations in simian virus 40. Proc. nat. Acad. Sci. (Wash.) 72, 989–993 (1975)
Smith, J.D.: Transcription and processing of transfer RNA precursors. Progr. Nucl. Acid Res. Mol. Biol. 16, 25–73 (1976)
Waldeck, W., Chowdhury, K., Gruss, P., Sauer, G.: Random cleavage of superhelical SV 40 DNA by S1 nuclease. Biochim. biophys. Acta (Amst.) 425, 157–167 (1976)
Wang, J.C.: Interactions between twisted DNAs and enzymes: The effects of superhelical turns. J. molec. Biol. 87, 797–816 (1974)
Weiss, B., Live, T.R., Richardson, C.C.: Enzymatic breakage and joining of DNA. V. End group labelling and analysis of DNA containing single strand breaks. J. biol. Chem. 243, 4530–4542 (1968)
Whitehouse, H.L.K., Hastings, P.J.: Analysis of genetic recombination on the polaron hybrid DNA model. Genet. Res. 6, 27–92 (1965)
Wiegand, R.C., Godson, G.N., Radding, C.M.: Specificity of the S1 nuclease from Aspergillus oryzae. J. biol. Chem. 250, 8848–8855 (1975)
Yamamoto, K.R., Alberts, B.M.: Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology 40, 734–744 (1970)
Zickler, H.: Genetische Untersuchungen an einen heteothallischen Askomyzeten (Bombardia lunata nov. spec.). Planta (Berl.) 22, 573–613 (1934)
Author information
Authors and Affiliations
Additional information
Communicated by F. Stahl
Rights and permissions
About this article
Cite this article
Pukkila, P.J. The recognition of mismatched base pairs in DNA by DNase I from Ustilago maydis . Molec. gen. Genet. 161, 245–250 (1978). https://doi.org/10.1007/BF00330997
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00330997