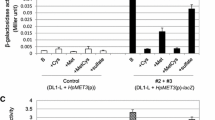
Summary
In yeast, as in other organisms, amino acid biosynthetic pathways share a common regulatory control. The manifestation of this control is that derepression of the enzymes belonging to several amino acid biosynthetic pathways follows amino acid starvation or tRNA discharging.
The arginine anabolic and catabolic pathways are, in addition, regulated specifically by arginine in opposite ways by common regulators. We have measured the mRNA levels for four genes subject to the general amino acid control: HIS4, ARG3, ARG4 and CPAII and compared them to the corresponding enzyme levels. Similarly we have measured the mRNA levels for two genes subject to the arginine specific regulation: ARG3 and CAR1, the former gene belongs to the arginine anabolic pathway and the latter to the arginine catabolic one.
HIS4, ARG4 and CPAII enzyme and messenger amounts are perfectly coordinated in all the conditions of general repression or derepression tested. However, arginine does not reduce the level of the ARG3 mRNA enough to explain the reduction of ornithine carbamoyltransferase activity nor does it increase the level of the CAR1 mRNA enough to explain the extent of induction of arginase. Coordination of enzyme and ARG3 mRNA is achieved only when the specific control is eliminated.
The half-lives of the ARG3 and CAR1 messengers are enhanced in mutants leading to constitutive expression of ornithine carbamoyltransferase and arginase.
These data suggest that the control that coordinates the synthesis of all the amino acids in the yeast cell operates at the level of transcription while the arginine specific regulatory mechanism seems to operate at a post-transcriptional level.
Similar content being viewed by others
References
Alwine CJ, Kemp JD, Parker B, Renart J, Stark RG, Wahl GM (1977) Detection of specific fragments of DNA by fractionation in gels and transfer to diazobenzylmethyl paper. Proc Natl Acad Sci USA 74:5350–5354
Béchet J, Grenson M, Wiame JM (1970) Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem 12:31–39
Bossinger J, Cooper TG (1977) Molecular events associated with induction of arginase in Saccharomyces cerevisiae. J Bacteriol 131:163–173
Carlson M, Botstein D (1982) Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of Yeast invertase. Cell 28:145–154
Carsiotis M, Lacy AM (1965) Increased activity of tryptophan biosynthetic enzymes in histidine mutants of Neurospora crassa. J Bacteriol 89:1472–1477
Crabeel M, Messenguy F, Lacroute F, Glansdorff N (1982) Cloning arg3, the gene for ornithine carbamoyltransferase from S. cerevisiae. Expression in E. coli requires secundary mutations. Production of plasmid β-lactamase in yeast. Proc Natl Acad Sci USA 78:5026–5030
Delforge J, Messenguy F, Wiame JM (1975) The regulation of arginine biosynthesis in Saccharomyces cerevisiae. The specificity of argR - mutations and the general control of amino acid biosynthesis. Eur J Biochem 57:231–239
Donahue T, Daves R, Lucchini G, Fink G (Jan 1983) A short nucleotide sequence required for regulation HIS4 by the general control system of yeast. Cell: (in press)
Dubois E, Grenson M, Wiame JM (1974) The participation of the anabolic glutamate dehydrogenase in the nitrogen catabolite repression of arginase in Saccharomyces cerevisiae. Eur J Biochem 48:603–616
Dubois E, Hiernaux D, Grenson M, Wiame JM (1978) Specific induction of catabolism and its relation to repression of biosynthesis in arginine metabolism of Saccharomyces cerevisiae. J Mol Biol 122:383–406
Dubois E, Messenguy F (1982) Cloning of two regulatory genes controlling the arginine metabolism in yeast. Arch Intern Physiol Biochim (in press)
Fink GR (1964) Gene enzyme relations in histidine biosynthesis in yeast. Science 146:525–527
Fraser RSS, Creanor J (1974) Rapid and selective inhibition of RNA synthesis in yeast by 8-hydroxyquinoline. Eur J Biochem 46:67–73
Hinnen A, Farabaugh P, Hilgen C, Fink GR, Friesen J (1979) Eucaryotic gene regulation. Aseel R, Maniatis T (eds) Academic Press NY, New York, p 43
Jacobs P, Jauniaux JC, Grenson M (1980) A cis-dominant regulatory mutation linked to the argB-argC cluster in Saccharomyces cerevisiae. J Mol Biol 139:691–704
Jauniaux JC, Dubois E, Vissers S, Crabeel M, Wiame JM (1982) Molecular cloning, DNA structure and RNA analysis of the arginase gene in Saccharomyces cerevisiae. A study of cisdominant regulatory mutations. Embo Journal, vol 1, n°9
Lowry OH, Rosebrough NJ, Farr L, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Mac Master G, Carmichael GG (1977) Analysis of single- and double stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci USA 74:4835–4838
Messenguy F, Wiame JM (1969) The control of ornithine carbamoyltransferase activity by arginase in Saccharomyces cerevisiae. FEBS Letters 3:47–49
Messenguy F, Penninckx M, Wiame JM (1971) Interaction between arginase and ornithine carbamoyltransferase in Saccharomyces cerevisiae. Eur J Biochem 22:277–286
Messenguy F, Delforge J (1976) Role of transfer ribonucleic acids in the regulation of several biosyntheses in Saccharomyces cerevisiae. Eur J Biochem 67:335–339
Messenguy F (1976) Regulation of arginine biosynthesis in Saccharomyces cerevisiae: isolation of a cis-dominant constitutive mutant for ornithine carbamoyltransferase synthesis. J Bacteriol 128:49–55
Messenguy F, Cooper TG (1977) Evidence that specific and general control of ornithine carbamoyltransferase production occurs at the level of transcription in Saccharomyces cerevisiae. J Bacteriol 130:1253–1261
Messenguy F (1979) Concerted repression of the synthesis of the arginine biosynthetic pathway by amino acids: a comparison between the regulatory mechanisms controlling amino acid biosyntheses in bacteria and yeast. Mol Gen Genet 169:85–95
Middelhoven WJ (1968) The derepression of arginase and of ornithine carbamoyltransferase in nitrogen starved baker's yeast. Biochim Biophys Acta 156:440–443
Piérard A, Messenguy F, Feller A, Hilger F (1979) Dual regulation of the synthesis of the arginine pathway carbamoylphosphate synthase of Saccharomyces cerevisiae by specific and general controls. Mol Gen Genet 174:163–171
Ramos F, Thuriaux P, Wiame JM, Béchet J (1970) Eur J Biochem 12:40–47
Rigby WJB, Dieckermann M, Rhodes C, Berg P (1977) Labelling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol 113:237–251
Schürch A, Miozzari J, Hütter R (1974) Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan sensitive mutants. J Bacteriol 117:1131–1140
Silverman SJ, Rose M, Botstein D, Fink GR (1982) Regulation of HIS4-lacZ fusions in Saccharomyces cerevisiae. Mol Cell Biol 2:1212–1219
St-John T, Davis R (1979) Isolation of galactose-inducible DNA sequences from Saccharomyces cerevisiae by differential plaque filter hybridization. Cell 16:443–452
Stephens JC, Artz SW, Ames BN (1975) Guanosine 5′-diphosphate 3′-diphosphate (ppGpp): positive effector for histidine operon transcription and general signal for amino acid deficiency. Proc Natl Acad Sci USA 72:4389–4393
Struhl K (1981) Deletion mapping of a eukaryotic promoter Proc Natl Acad Sci USA 78:4461–4465
Waldron C, Lacroute F (1975) Effect of growth rate on the amounts of ribosomal and transfer ribonucleic ends in yeast. J Bacteriol 122:855–865
Wiame JM (1971) The regulation of arginine metabolism in Saccharomyces cerevisiae. Exclusion mechanisms. In: Horecker BL, Stadtman ER (eds) Current topics in cellular regulation, vol 4. Academic Press, New York, pp 1–38
Winckler ME, Roth DJ, Hartman PE (1978) Promoter and attenuator-related metabolic regulation of Salmonella typhimurium histidine operon. J Bacteriol 133:830–843
Wolfner M, Yep D, Messenguy F, Fink GR (1975) Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol 96:273–290
Author information
Authors and Affiliations
Additional information
Communicated by G. Fink
Rights and permissions
About this article
Cite this article
Messenguy, F., Dubois, E. Participation of transcriptional and post-transcriptional regulatory mechanisms in the control of arginine metabolism in yeast. Molec Gen Genet 189, 148–156 (1983). https://doi.org/10.1007/BF00326068
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00326068