Abstract
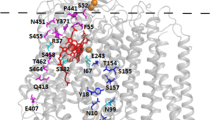
Three respiratory-deficient mutants of cytochrome oxidase subunit I in the yeast mitochondrion have been sequenced. They are located in, or near, transmembrane segment VI, the catalytic core of the enzyme. Respiratory-competent revertants have been selected and studied. The mutant V244M was found to revert at the same site in valine (wild-type), isoleucine or threonine. The revertants of the mutant G251R were of three types: glycine (wild-type), serine and threonine at position 251. A search for second-site mutations was carried out but none were found. Among 60 revertants tested, the mutant K265M was found to revert only to the wild-type allele.
Similar content being viewed by others
References
Babcock GT, Wikström M (1992) Oxygen activation and the conservation of energy in cell respiration. Nature 356:301–309
Bairoch A, Boeckmann B (1993) The SWISS-PROT protein sequence data bank, recent developments. Nucleic Acids Res 21:3093–3096
Briquet M, Goffeau A (1981) Modification of the spectral properties of cytochrome b in mutants of Saccharomyces cerevisiae resistant to 3-(3,4-dichlorophenyl)-1,1-dimethylurea. Eur J Biolchem 117:333–339
Brown S, Moody AJ, Mitchell R, Rich PR (1993) Binuclear centre structure of terminal protonmotive oxidases. FEBS Lett 316:216–223
Carpenter C, Simon A (1990) Simplified RNA sequencing using dideoxy chain-termination. Biotechniques 8:26–27
Claisse M, Aubert G, Clavilier L, Slonimski PP (1970) Méthode d'estimation de la concentration des cytochromes dans les cellules entières de levure. Eur J Biochem 16:430–438
Cooper CE, Nicholls P, Freedman JA (1991) Cytochrome-c oxidase — structure, function, and membrane topology of the polypeptide subunits. Biochem Cell Biol 69:586–607
Cooper CE, Jünemann S, Ioannidis N, Wrigglesworth JM (1993) Slow (resting) forms of mitochondrial cytochrome c oxidase consist of two kinetically distinct conformations of the binuclear CuB/a 3 centre — relevance to the mechanism of proton translocation. Biochim Biophys Acta 1144:149–160
Crowe JS, Cooper HJ, Smith MA, Parker D, Gewert D (1991) Improved cloning efficiency of polymerase chain reaction (PCR) products after proteinase K digestion. Nucleic Acids Res 19:184
di Rago JP, Colson A (1988) Molecular basis for resistance to antimycine and diuron, Q-cycle inhibitors acting at the Qi site in the mitochondrial ubiquinol-cytochrome c reductase in Saccharomyces cerevisiae. J Biol Chem 263:12564–12570
di Rago JP, Netter P, Slonimski PP (1990a) Intragenic suppressors reveal long-distance interactions between inactivating and reactivating amino-acid replacements generating three-dimensional constraints in the structure of mitochondrial cytochrome b. J Biol Chem 265:15750–15757
di Rago JP, Netter P, Slonimski PP (1990b) Pseudo-wild-type revertants from inactive apocytochrome b mutants as a tool for the analysis of the structure-function relationships of the mitochondrial ubiquinol-cytochrome c reductase of Saccharomyces cerevisiae. J Biol Chem 265:3332–3339
Fox TD (1987) Natural variation in the genetic code. Annu Rev Genet 21:67–91
Harris SL, Perlin DS, Seto-Young D, Haber JE (1991) Evidence for coupling between membrane and cytoplasmic domains of the yeast plasma membrane H+-ATPase. Analysis of revertants of pma-105. J Biol Chem 266:24439–24445
Huttenhofer A, Weiss-Brummer B, Dirheimer G, Martin RP (1990) A novel type of +1 frameshift suppressor: a base substitution in the anticodon stem of a yeast mitochondrial serine-tRNA causes frameshift suppression. EMBO J 9:551–558
Kadenbach B, Stroh A, Huther F-J, Reinman A, Steyerding D (1991) Evolutionary aspects of cytochrome c oxidase. J Bioenerg Biomembr 23:321–334
Kotylak Z, Slonimski PP (1977) Mitochondrial mutants isolated by a new screening method based upon the use of the nuclear mutation op1. In: Bandlow W, Schweyen RJ, Wolf K, Kaudewitz F (eds) Mitochondria 1977: genetics and biogenesis of mitochondria. Walter de Gruyter, Berlin, pp 83–89
Lancashire W, Mattoon J (1979) Cytoduction: a tool for mitochondrial genetic studies in yeast. Mol Gen Genet 170:333–344
McCabe PC (1990) Production of single-stranded DNA by asymetric PCR. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols. Academic Press, New York, pp 76–83
Meunier B, Colson AM (1993) Insight into the interactions between subunits I and II of the cytochrome c oxidase of the yeast Saccharomyces cerevisiae by means of extragenic complementation. FEBS Lett 335:338–340
Meunier B, Coster F, Lemarre P, Colson AM (1993) Insight into topological and functional relationships of cytochrome-c oxidase subunit-I of Saccharomyces cerevisiae by means of intragenic complementation. FEBS Lett 321:159–162
Minagawa J, Mogi Gennis R, Anraku Y (1992) Identification of heme and copper ligands in subunit-I of the cytochrome-bo complex in Escherichia coli. J Biol Chem 267:2096–2104
Nelson DR, Douglas MG (1993) Function-based mapping of the yeast mitochondrial ADP/ATP translocator by selection for second-site revertants. J Mol Biol 230:1171–1182
Netter P, Carignani G, Jacq C, Groudinsky O, Clavillier L, Slonimski PP (1982) The cytochrome oxidase subunit-I split gene in Saccharomyces cerevisiae: genetic and physical studies of the mtDNA segment encompassing the ‘cytochrome b-homologuous’ intron. Mol Gen Genet 188:551–559
Netter P, Robineau S, Sirandpugnet P, Fauvarque MO (1992) The unusual reversion properties of a mitochondrial mutation in the structural gene of subunit I of cytochrome oxidase of Saccharomyces cerevisiae reveal a probable bistidine ligand of the redox center. Curr Genet 21:147–151
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, New York
Saraste M (1990) Structural features of cytochrome oxidase. Quart Rev Biophys 23:331–366
Shen Z, Fox TD (1989) Substitution of an invariant nucleotide at the base of the highly-conserved ‘530-loop’ of 15s rRNA causes suppression of yeast mitochondrial ochre mutations. Nucleic Acids Res 17:4535–4538
Author information
Authors and Affiliations
Additional information
Communicated by K. Wolf
Rights and permissions
About this article
Cite this article
Lemarre, P., Robineau, S., Colson, AM. et al. Sequence analysis of three deficient mutants of cytochrome oxidase subunit I of Saccharomyces cerevisiae and their revertants. Curr Genet 26, 546–552 (1994). https://doi.org/10.1007/BF00309948
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00309948