Abstract
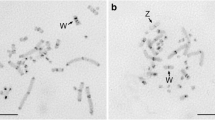
DNA satellites were isolated from three balenopterid species, viz. the minke, sei, and fin whales. In each of them at least two DNA satellites were recognizable with buoyant densities in neutral CsCl of ρ=1.702/1.703 and ρ=1.710/1.711, respectively. cRNAs from each satellite group were used for filter and in situ hybridisations. Homo- and heterologous DNA-cRNA hybrids within each satellite group yielded virtually identical melting curve profiles showing conservation of at least a considerable part of the DNA satellite sequences. There was no evident sequence homology between the ρ=1.702/1.703 and the ρ=1.710/1.711 satellites by filter hybridisation. — The in situ hybridisation showed that in each species the ρ=1.702/1.703 satellite was located in centromeric-paracentromeric C-bands in a few pairs, whereas the ρ=1.710/1.711 satellite was located in terminal C-bands throughout the karyotypes. — The data on the whale DNA satellites indicate that the quantitative evolution of the satellite DNA sequences preceded species divergence of the balenopterids and that the satellite sequences have remained relatively unaltered since the divergence took place. The function of satellite DNA is considered to imply the introduction of both chromosomal and genic polymorphisms and thus being of great importance in speciation. Based upon these concepts a model is postulated for the function of satellite DNA. According to this model at meiotic pairing euchromatin-heterochromatin overlapping between homologous chromosomes is considered to be of a general occurrence. This overlapping is presumed to be accentuated by the size heteromorphism frequently observed between homologous heterochromatic segments (C-bands). In the region of such euchromatin-heterochromatin overlapping, crossing-over would be excluded. The overlapping is suggested to be rectified progressively in the chromosome arms, leaving unaffected crossing-over distant to the euchromatin-heterochromatin junctions. The consequence of this will be that genes in the proximity of the junctions are collectively inherited and selected, whereas genes distant to the heterochromatin will be independently assorted and selected.
Similar content being viewed by others
References
Árnason, Ú.: The role of chromosomal rearrangement in mammalian speciation with special reference to Cetacea and Pinnipedia. Hereditas (Lund) 70, 112–118 (1972)
Árnason, Ú.: Comparative chromosome studies in Cetacea, Hereditas (Lund) 77, 1–36 (1974a)
Árnason, Ú.: Comparative chromosome studies in Pinnipedia. Hereditas (Lund) 76, 179–226 (1974b)
Árnason, Ú.: Phylogeny and speciation in Pinnipedia and Cetacea — A cytogenetic study. Thesis, University of Lund (1974b)
Árnason, Ú.: The relationship between the four principal pinniped karyotypes. Hereditas (Lund) 87, 227–242 (1977)
Árnason, Ú., Benirschke, K., Nichols, W.W., Mead, J.G.: Banded karyotypes of three whales: Mesoplodon europaeus, M. carlhubbsi and Balaenoptera acutorostrata. Hereditas (Lund) 87, 189–200 (1977)
Birnstiel, M.L., Sells, B.H., Purdom, I.F.: Kinetic complexity of RNA molecules. J. molec. Biol. 63, 21–39 (1972)
Birnstiel, M.L., Spiers, J., Purdom, I.F., Jones, K., Loening, U.E.: Properties and composition of the isolated ribosomal DNA satellite of Xenopus laevis. Nature (Lond.) 219, 454–463 (1968)
Callan, H.G., Lloyd, L.: Lampbrush chromosomes. In: New Approaches in cell biology (P.M.B. Walker, ed.), pp. 23–46. New York: Academic Press 1960
Gillespie, D., Spiegelman, S.: A quantitative assay for DNA-RNA hybrids with DNA immobilised on a membrane. J. molec. Biol. 12, 829–842 (1965)
Jensen, R.H., Davidson, N.: Spectrophotometric, potentiometric and density gradient ultracentrifugation studies of the binding of silver ion by DNA. Biopolymers 4, 17–32 (1966)
Jones, K.W.: Chromosomal and nuclear location of mouse satellite DNA in individual cells. Nature (Lond.) 225, 912–915 (1970)
Jones, K.W.: In situ hybridisation. In: New techniques in biophysics and cell biology (R.H. Pain and B.J. Smith, eds.), Vol. 1, pp. 29–66. London: J. Wiley and Sons 1973
Jones, K.W.: Repetitive DNA and primate evolution. In: Chromosomes in biology and medicine. Vol. 1, The molecular structure of human chromosomes (J.J. Yunis, ed.), pp. 295–326. New York: Academic Press Inc. 1977
Lubs, H.A., Kimberling, W.J., Hecht, F., Patil, S.R., Brown, J., Gerald, P., Summitt, R.L.: Racial differences in the frequency of Q and C chromosomal heteromorphisms. Nature (Lond.) 268, 631–633 (1977)
Macgregor, H.C., Andrews, G.: The arrangement and transcription of “middle repetitive” DNA sequences on lampbrush chromosomes of Triturus. Chromosoma (Berl.) 63, 109–126 (1977)
Marmur, J.: A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. molec. Biol. 3, 208–218 (1961)
Merker, A.: The cytogenetic effect of heterochromatin in hexaploid triticale. Hereditas (Lund) 83, 215–222 (1976)
Miklos, G.L.G., Nankivell, R.N.: Telomeric satellite DNA functions in regulating recombination, Chromosoma (Berl.) 56, 143–167 (1976)
Moar, M.H., Purdom, I.F., Jones, K.W.: Influence of temperature on the detectibility and chromosomal distribution of specific DNA sequences by in situ hybridisation. Chromosoma (Berl.) 53, 345–359 (1975)
Müller, H., Klinger, H.P.: Chromosome polymorphism in a human newborn population—Part 1. In: Chromosomes today 5, 249–260 (1974)
Nankivell, R.N.: Karyotype differences in the crenaticeps-group of Atractomorpha (Orthoptera, Acridoidea, Pyrgomorphidae). Chromosoma (Berl.) 56, 127–142 (1976)
Pardue, M.L., Gall, J.G.: Chromosomal localisation of mouse satellite DNA. Science 168, 1356–1358 (1970)
Pathak, S., Stock, A.D.: The X chromosomes of mammals: Karyological homology as revealed by banding techniques. Genetics 78, 703–714 (1974)
Singh, L., Purdom, I.F., Jones, K.W.: Satellite DNA and evolution of sex chromosomes. Chromosoma (Berl.) 59, 43–62 (1976)
Singh, L., Purdom, I.F., Jones, K.W.: Effect of different denaturing agents on the detectability of specific DNA sequences of various base compositions by in situ hybridisation. Chromosoma (Berl.) 60, 377–389 (1977)
Sumner, A.T.: A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 75, 304–306 (1972)
Thomas, J.B., Kaltsikes, P.J.: A possible effect of heterochromatin on chromosome pairing. Proc. nat. Acad. Sci. (Wash.) 71, 2787–2790 (1974)
Yamamoto, M, Miklos, G.L.G.: Genetic dissection of heterochromatin in Drosophila: The role of basal X heterochromatin in meiotic sex chromosome behaviour. Chromosoma (Berl.) 60, 283–296 (1977)
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Árnason, Ú., Purdom, I.F. & Jones, K.W. Conservation and chromosomal localization of DNA satellites in balenopterid whales. Chromosoma 66, 141–159 (1978). https://doi.org/10.1007/BF00295136
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00295136