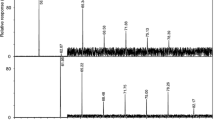
Summary
The cellular long-chain fatty acids of 69 strains of yeasts, representing 29 species associated with the brewing industry, were extracted by saponification and analyzed asmethyl esters by gas chromatography. The strains were characterized by the presence of palmitic, palmitoleic, stearic, oleic, linoleic and linolenic acid as the major fatty acids. The strains were divided into six groups on the basis of their fatty acid content. With this method it was possible to differentiate between the yeasts on species and, in some instances, on strain level.
Similar content being viewed by others
References
Abel K, Schmertzing H, Peterson JI (1963) Classification of microorganisms by analysis of chemical composition. I. Feasibility of utilizing gas chromatography. J Bacteriol 85:1039–1044
Baptist JN, Kurtzman CP (1976) Comparative enzyme patterns in Cryptococcus laurentii and its taxonomic varieties. Mycologia 68:1195–1203
Campbell I (1972) Numerical analysis of the genera Saccharomyces and Kluyveromyces. J Gen Microbiol 73:279–301
Campbell I (1973) Comparative identification of yeasts of the genus Saccharomyces. J Gen Microbiol 77:127–135
Campbell I (1974) Methods of numerical taxonomy of various genera of yeasts. Adv Appl Microbiol 17:135–155
Cottrell M, Kock JLF, Lategan PM, Botes PJ, Britz TJ (1985) The long-chain fatty acid composition of species representing the genus Kluyveromyces. FEMS Microbiol Lett 30:373–376
Dees SB, Moss CW (1975) Cellular fatty acids Alcaligenes and Pseudomonas species isolated from clinical specimens. J Clin Microbiol 1:414–419
Gorin PAJ, Spencer JFT (1970) Proton magnetic resonance spectroscopy — an aid in identification and chemotaxonomy of yeasts. Adv Appl Microbiol 13:25–89
Kock JLF, Lategan PM, Botes PJ, Viljoen BC (1985a) Developing a rapid statistical identification process for different yeast species. J Microbiol Meth 4:147–154
Kock JLF, Botes PJ, Erasmus SC, Lategan PM (1985b) A rapid method to differentiate between four species of the Endomycete. J Gen Microbiol 31:3393–3396
Kock JLF, Cottrell M, Lategan PM (1986a) A rapid method to differentiate between five species of the genus Saccharomyces. Appl Microbiol Biotechnol 23:499–501
Kock JLF, Erasmus SC, Lategan PM (1986b) A rapid method to identify some species of the Dipodascaceae and Endomycetaceae. J Microbiol Meth (In Press)
Kurtzman CP, Smiley MJ, Baker FL (1972) Scanning electron microscopy of ascospores of Schwanniomyces. J Bacteriol 112:1380–1382
Kurtzman CP, Smiley MJ (1974) A taxonomic evaluation of the round-spored species of Pichia. In: Klaushöfer H, Sleytr UB (eds) Proceedings of the Fourth International Symposium on Yeasts. Vienna, Austria
Kurtzman CP, Smiley MJ, Bakter FL (1975) Scanning electron-microscopy of ascospores of Debaryomyces and Saccharomyces. Mycopathol Mycol Appl 55:29–34
Lodder J, Sloof WC, Kreger-van Rij NJW (1958) The classification of yeasts. In: Coock AH (ed) The chemistry and biology of yeasts. Academic Press, New York
Price CW, Fuson GB, Phaff HJ (1978) Genome comparison in yeast systematics: delimination of species within the genera Schwanniomyces, Saccharomyces, Debaryomyces and Pichia. Micr Rev 42:161–193
Scheda R, Yarrow D (1966) The instability of physiological properties used as criteria in the taxonomy of yeasts. Arch Microbiol 55:209–225
Tsuchiya T, Fukuzawa Y, Taguchi M, Nakase T, Shinoda T (1974) Serological aspects on yeast classification. Mycopathol Mycol Appl 53:77–91
Van der Walt JP (1970) Genus 13. Kluyveromyces van der Walt emend. van der Walt. In: Lodder J (ed) The Yeasts: a taxonomic study. 2nd revised and enlarged edition, North Holland Publishing Company, Amsterdam and London
Viljoen BC, Kock JLF, Lategan PM (1986) Long-chain fatty acid composition of selected genera of yeasts belonging to the Endomycetales. Antonie van Leeuwenhoek 52:45–51
Yamada Y, Arimoto M, Kondo K (1973) Coenzyme Q system in the classification of the ascosporogenous yeast genus Schizosaccharomyces and the yeast-like genus Endomyces. J Gen Appl Microbiol 19:353–358
Yamada Y, Arimoto M, Kondo K (1976) Coenzyme Q system in the classification of apiculate yeast in the genera Nadsonia, Saccharomycodes, Hanseniaspora, Kloekera and Wickerhamia. J Gen Appl Microbiol 22:293–299
Yamada Y, Arimoto M, Kondo K (1977) Coenzyme Q system in the classification of some ascosporogenous yeast genera in the families Saccharomycetaceae and Spermophthoraceae. Antonie van Leeuwenhoek 43:65–72
Yamada Y, Kondo K (1972) Taxonomic significance of the coenzyme Q system in yeasts and yeast-like fungi (2). In: Terui G (ed) Fermentation technology today proceedings of the Fourth International Fermentation Symposium. Society of Fermentation Technology. Osaka, Japan
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Oosthuizen, A., Kock, J.L.F., Botes, P.J. et al. The long-chain fatty acid composition of yeasts used in the brewing industry. Appl Microbiol Biotechnol 26, 55–60 (1987). https://doi.org/10.1007/BF00282149
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00282149