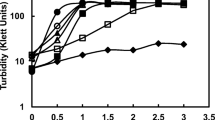
Summary
Spontaneous chlorate-resistant (CR) mutants have been isolated from Chlamydomonas reinhardtii wildtype strains. Most of them, 244, were able to grow on nitrate minimal medium, but 23 were not. Genetic and in vivo complementation analyses of this latter group of mutants indicated that they were defective either at the regulatory locus nit-2, or at the nitrate reductase (NR) locus nit-1, or at very closely linked loci. Some of these nit-1 or nit-2 mutants were also defective in pathways not directly related to nitrate assimilation, such as those of amino acids and purines. Chlorate treatment of wild-type cells resulted in both a decrease in cell survival and an increase in mutant cells resistant to a number of different chemicals (chlorate, methylammonium, sulphanilamide, arsenate, and streptomycin). The toxic and mutagenic effects of chlorate in minimal medium were not found when cells were grown either in darkness or in the presence of ammonium, conditions under which nitrate uptake is drastically inhibited. Chlorate was also able to induce reversion of nit − mutants of C. reinhardtii, but failed to produce His + revertants or Arar mutants in the BA-13 strain of Salmonella typhimurium. In contrast, chlorate treatment induced mutagenesis in strain E1F1 of the phototrophic bacterium Rhodobacter capsulatus. Genetic analyses of nitrate reductase-deficient CR mutants of C. reinhardtii revealed two types of CR, to low (1.5 mM) and high (15 mM) chlorate concentrations. These two traits were recessive in heterozygous diploids and segregated in genetic crosses independently of each other and of the nit-1 and nit-2 loci. Three her loci and four lcr loci mediating resistance to high (HC) and low (LC) concentrations of chlorate were identified. Mutations at the nit-2 locus, and deletions of a putative locus for nitrate transport were always epistatic to mutations responsible for resistance to either LC or HC. In both nit + and nit − chlorate-sensitive (CS) strains, nitrate and nitrite gave protection from the toxic effect of chlorate. Our data indicate that in C. reinhardtii chlorate toxicity is primarily dependent on the nitrate transport system and independent of the existence of an active NR enzyme. At least seven loci unrelated to the nitrate assimilation pathway and mediating CR are thought to control indirectly the efficiency of the nitrate transporter for chlorate transport. In addition, chlorate appears to be a mutagen capable of inducing a wide range of mutations unrelated to the nitrate assimilation pathway.
Similar content being viewed by others
References
Aberg B (1947) On the mechanism of the toxic action of chlorates and some related substances upon young wheat plants. Ann Kungl Lantbrukshögskol 15:37–107
Aguilar MR, Prieto R, Cárdenas J, Fernández E (1992) nit-7: a new locus for molybdopterin cofactor biosynthesis in the green alga Chlamydomonas reinhardtii. Plant Physiol 98:395–398
Arnon DI (1949) Copper enzymes in isolated chloroplast. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Caboche M, Rouzé P (1990) Nitrate reductase: a target for molecular and cellular studies in higher plants. Trends Genet 6:187–192
Cove DJ (1976) Chlorate toxicity in Aspergillus nidulans. Studies of mutants altered in nitrate assimilation. Mol Gen Genet 146:147–159
Deane-Drummond CE (1984) Nitrate transport into Chara coralina cells using 36ClO −3 as an analogue for nitrate. I. Interaction between 36ClO −3 and NO −3 and characterization of 36ClO −3 / NO −3 Nitrate transport into Chara coralina influx. J Exp Bot 35:1289–1298
Deane-Drummond CE (1988) Characteristics of 36ClO −3 uptake into Pisum sativum seedlings. Limitations and uses of 36ClO −3 as an analogue for NO −3 . Plant Sci 42:99–108
Deane-Drummond CE, Glass ADM (1983) Short term studies of nitrate uptake into barley plants using ion-specific electrodes and 36ClO −3 . I. Control of net uptake by NO −3 efflux. Plant Physiol 73:100–104
Doddema H, Telkamp GP (1979) Uptake of nitrate by mutants of Arabidopsis thaliana disturbed in uptake or reduction of nitrate. Physiol Plant 45:332–338
Dorado G, Pueyo C (1988) l-Arabinose resistance test with Salmonella typhimurium as a primary tool for carcinogen screening. Cancer Res 48:907–912
Ebersold WT (1967) Chlamydomonas reinhardtii: heterozygous diploid strains. Science 157:447–449
Fernández E, Matagne RF (1984) Genetic analysis of nitrate reductase-deficient mutants in Chlamydomonas reinhardtii. Curr Genet 8:635–640
Fernández E, Matagne RF (1986) In vivo complementation analysis of nitrate reductase-deficient mutants in Chlamydomonas reinhardtii. Curr Genet 10:397–403
Fernández E, Schnell RA, Ranum LPW, Hussey SC, Silflow CD, Lefebvre PA (1989) Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 86:6449–6453
Fischer P, Klein U (1988) Localization of nitrogen-assimilating enzymes in the chloroplast of Chlamydomonas reinhardtii. Plant Physiol 88:947–952
Florencio FJ, Vega JM (1983) Utilization of nitrate, nitrite and ammonium by Chlamydomonas reinhardtii. Photoproduction of ammonium. Planta 158:288–293
Galván A, Cárdenas J, Fernández E (1992) Nitrate reductase regulates expression of nitrite uptake and nitrite reductase activities in Chlamydomonas reinhardtii. Plant Physiol 98:422–426
Garrett RH (1972) The induction of nitrite reductase in Neurospora crassa. Biochim Biophys Acta 264:481–489
Guy M, Zabala G, Filner P (1988) The kinetics of chlorate uptake by XD tobacco cells. Plant Physiol 86:817–821
Harris E (1989) The Chlamydomonas sourcebook. Academic Press, New York
Hofstra JJ (1977) Chlorate toxicity and nitrate reductase activity in tomato plants. Physiol Plant 41:65–69
Lewis CM, Fincham JRS (1970) Genetics of nitrate reductase in Ustilago maydis. Genet Res 16:151–163
Levine RP, Ebersold WT (1960) The genetics and cytology of Chlamydomonas. Annu Rev Microbiol 14:197–216
Liljeström S, Aberg B (1966) Studies on the mechanism of chlorate toxicity. Ann Kungl Lantbrukshögskol 32:93–107
López-Ruíz A, Verbelen JP, Roldán JM, Díez J (1985) Nitrate reductase of green algae is located in the pyrenoid. Plant Physiol 79:1006–1010
Muñoz-Blanco J, Hidalgo-Martínez J, Cárdenas J (1990) Extracellular deamination of l-amino acids by Chlamydomonas reinhardtii cells. Planta 182:194–198
Müller AJ, Grate R (1978) Isolation and characterization of cell lines of Nicotiana tabacum lacking nitrate reductase. Mol Gen Genet 161:67–76
Nussaume L, Vincentz M, Caboche M (1991) Constitutive nitrate reductase: a dominant conditional marker for plant genetics. Plant J 1:267–274
Okabe Y, Okada M (1990) Nitrate reductase activity and nitrite in native pyrenoids purified from the green alga Bryopsis maxima. Plant Cell Physiol 31:429–432
Oostindiër-Braaksma FJ, Feenstra WJ (1973) Isolation and characterization of chlorate-resistant mutants of Arabidopsis thaliana. Mutat Res 19:175–185
Pichinoty F (1966) Proprietés, régulation et fonctions physiologiques des nitrate-réductase bacteriennes A et B. Bull Soc Fr Physiol Veg 12:97–104
Quesada A (1992) Mecanismos moleculares de la asimilación de nitrato en algas verdes. Ph D Dissertation. University of Córdoba (Spain)
Ruíz-Rubio M, Hera C, Pueyo C (1984) Comparison of a forward and a reverse mutation assay in Salmonella typhimurium measuring l-arabinose resistance and histidine prototrophy. EMBO J 3:1435–1440
Solomonson LP, Vennesland B (1972) Nitrate reductase and chlorate toxicity in Chlorella vulgaris Beijerinck. Plant Physiol 50:421–424
Sosa FM, Ortega T, Barea JL (1978) Mutants from Chlamydomonas reinhardtii affected on their nitrate assimilation capability. Plant Sci Lett 11:51–58
Tomsett AB, Garrett RH (1980) The isolation and characterization of mutants defective in nitrate assimilation in Neurospora crassa. Genetics 93:649–660
Vaughn K, Campbell WH (1988) Immuno-gold location of nitrate reductase in maize leaves. Plant Physiol 88:1354–1357
Vega JM (1972) NADH-nitrate réductase de Chlorella. Nouvelle contribution à l'étude de ses propriétés. Physiol Vég 10:637–652
Weaver PF, Wall JD, Gest H (1975) Characterization of Rhodopseudomonas capsulata Arch Microbiol 105:207–216
Wray JL (1986) The molecular genetics of higher plant nitrate assimilation. In: Blonstein AD, King PJ (eds) A genetic approach to plant biochemistry. Springer, New York, pp 101–157
Author information
Authors and Affiliations
Additional information
Communicated by R. Herrmann
Rights and permissions
About this article
Cite this article
Prieto, R., Fernández, E. Toxicity of and mutagenesis by chlorate are independent of nitrate reductase activity in Chlamydomonas reinhardtii . Molec. Gen. Genet. 237, 429–438 (1993). https://doi.org/10.1007/BF00279448
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00279448