Abstract
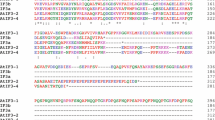
The positive-acting global sulfur regulatory protein, CYS3, of Neurospora crassa turns on the expression of a family of unlinked structural genes that encode enzymes of sulfur catabolism. CYS3 contains a leucine zipper and an adjacent basic region (b-zip), which together constitute a bipartite sequence-specific DNA-binding domain. Specific anti-CYS3 antibodies detected a protein of the expected size in nuclear extracts of wild-type Neurospora under conditions in which the sulfur circuit is activated. The CYS3 protein was not observed in cys-3 mutants. Nuclear extracts of wild type, but not cys-3 mutants, also showed specific DNA-binding activity identical to that obtained with a CYS3 protein expressed in Escherichia coli. A truncated CYS3 protein that contains primarily the b-zip domain binds to DNA with high specificity and affinity in vitro, yet fails to activate gene expression in vivo, and instead inhibits the function of the wild-type CYS3 protein. Amino-terminal, carboxy-terminal, and internal deletions as well as alanine scanning mutagenesis were employed to identify regions of the CYS3 protein that are required for its trans-activation function. Regions of CYS3 carboxy terminal to the b-zip motif are not completely essential for function although loss of an alanine-rich region results in decreased activity. All deletions amino terminal to the b-zip motif led to a complete loss of CYS3 function. Alanine scanning mutagenesis demonstrated that an unusual proline-rich domain of CYS3 appears to be very important for function and is presumed to constitute an activation domain. It is concluded that CYS3 displays nuclear localization and positive autogenous control in Neurospora and functions as a trans-acting DNA-binding protein.
Similar content being viewed by others
References
Burton EG, Metzenberg RL (1971) Novel mutation causing derepression of several enzymes of sulfur metabolism in Neurospora crassa. J Bacteriol 109:140–151
Courey AJ, Tjian R (1988) Analysis of SP1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55:887–898
Comai L, Tanese N, Tjian R (1992) The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68:965–976
Desplan C, Theis J, O'Farrel PH (1988) The sequence specificity of homeodomain-DNA interaction. Cell 54:1081–1090
Dynlacht BD, Hoey T, Tjian R (1991) Isolation of coactivators associated with TATA-binding protein that mediate transcriptional activation. Cell 66:563–576
Ebbole D, Sachs MS (1990) A rapid -and simple method for isolation of Neurospora crassa homokaryons using microconidia. Neurospora Newslett 37:17–18
Fu YH, Marzluf GA (1990) Cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. J Biol Chem 265:11942–11947
Fu YH, Paietta JV, Mannix DG, Marzluf GA (1989) Cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a protein with a putative leucine zipper DNA-binding element. Mol Cell Biol 9:1120–1127
Fu YH, Lee HJ, Young JL, Jarai G, Marzluf GA (1990) Nitrogen and sulfur regulatory circuits of Neurospora. In: Davidson EH, Ruderman JV, Posakony JW (ds) Developmental Biology. Wiley-Liss, pp 319–335
Godowski PJ, Picard D, Yamamoto KR (1988) Signal transduction and transcriptional regulation by glucocorticoid receptor-lexA fusion proteins. Science 241:812–816
Hautala JA, Conner BH, Jacobson JW, Patel GL, Giles NH (1977) Isolation and characterization of nucleic from Neurospora crassa. J Bacteriol 130:704–713
Hope IA, Struhl K (1986) Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell 46:885–894
Hope IA, Mahadevan S, Struhl K (1988) Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature 333:635–640
Kanaan MN, Marzluf GA (1991) Mutational analysis of the DNA-binding domain of the CYS3 regulatory protein of Neurospora crassa. Mol Cell Biol 9:4356–4362
Kanaan MN, Fu YH, Marzluf GA (1992) The DNA-binding domain of the Cys-3 regulatory protein of Neurospora crassa is bipartite. Biochemistry 31:3197–3203
Ketter JS, Marzluf GA (1988) Molecular cloning and analysis of the regulation of cys-14, a structural gene of the sulfur regulatory circuit of Neurospora crassa. Mol Cell Biol 8:1504–1508
Kunkel TA (1985) Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA 82:488–492
Ma J, Ptashne M (1987) Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48:847–853
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Marzluf GA, Metzenberg RL (1968) Positive control by the cys-3 locus in regulation of sulfur metabolism in Neurospora crassa. J Mol Biol 33:423–437
Mermond N, O'Neill EA, Kelly TJ, Tjian R (1989) The proline-rich transcriptional activator of CTF/NF-1 is distinct from the replication and DNA-binding domain. Cell 58:741–753
Metzenberg RL, Parson JW (1966) Altered repression of some enzymes of sulfur utilization in a temperature-conditional lethal mutant of Neurospora. Proc Natl Acad Sci USA 55:629–635
Paietta JV (1989) Molecular cloning and regulatory analysis of the arylsulfatase structural gene of Neurospora crassa. Mol Cell Biol 9:3630–3637
Paietta JV (1990) Molecular cloning and analysis of the scon-2 negative regulatory gene of Neurospora crassa. Mol Cell Biol 10:5207–5214
Paietta JV (1992) Production of the CYS3 regulator, a bZIP DNA-binding protein, is sufficient to induce sulfur gene expression in Neurospora crassa. Mol Cell Biol 12:1568–1577
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Author information
Authors and Affiliations
Additional information
Communicated by C. van den Hondel
Rights and permissions
About this article
Cite this article
Kanaan, M.N., Marzluf, G.A. The positive-acting sulfur regulatory protein CYS3 of Neurospora crassa: nuclear localization, autogenous control, and regions required for transcriptional activation. Molec. Gen. Genet. 239, 334–344 (1993). https://doi.org/10.1007/BF00276931
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00276931