Summary
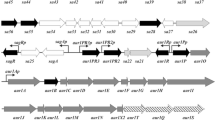
The gene encoding the Neisseria lactamica III DNA methyltransferase (M.NlaIII) which recognizes the sequence CATG has been cloned and expressed in Escherichia coli. DNA sequencing of a 3.125 kb EcoR1-Pst1 fragment localizes the M.NlaIII gene to a 334 codon open reading frame (ORF) and identifies, 468 by downstream, a second ORF of 313 amino acids, which is referred to as M.NlaX. Both proteins are detectable in the E. coli coupled in vitro transcription-translation system; they are apparently expressed from separate N. lactamica promoters. The N-terminal half of the previously characterized M.FokI, which methylates adenine in one of the DNA strands with its asymmetric recognition sequence (GGATG), is found to have 41% sequence identity and a further 11.7% sequence similarity with M.NlaIII. Among the conserved amino acids is the well-known DPPY sequence motif. With one exception, analysis of the nucleotides coding for the DP dipeptide in all known DPPY sequences shows the presence of an inherent DNA adenine methylation (dam) recognition site of GATC. A low level of expression of M.NlaX in E. coli prevents the elucidation of its sequence recognition specificity. Sequence analysis of M.NlaX shows that it is closely related to the group of monospecific 5-methylcytosine DNA methyltransferases (M.EcoRII, Dcm, M.HpaII and M.HhaI) which all have a modified cytosine at the second position of the recognition sequences. Both M.EcoRII and Dcm amino acid sequences are about 50% identical with M.NlaX; a considerable degree of sequence identity is found in the so-called variable region which is believed to be responsible for sequence recognition specificity. M.NlaX is probably the counterpart to the E. coli Dcm in N. lactamica.
Similar content being viewed by others
References
Barras F, Marinus MG (1989) The great GATC: DNA methylation in E. coli. Trends Genet 5:139–143
Bhagwat AS, Sohail A, Roberts RJ (1986) Cloning and characterization of the dcm locus of Escherichia coli K-12. J Bacteriol 166:751–755
Bickle TA (1987) DNA restriction and modification systems. In: Neidhardt FC (ed) Escherichia coli and Salmonella typhimurium, Cellular and molecular biology, vol. 1. American Society for microbiology, Washington DC, pp 692–696
Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heynecker HL, Boyer HW, Crosa JH, Falkow S (1977) Construction and characterization of new cloning vehicles II. A multipurpose cloning system. Gene 2:95–113
Bougueleret L, Schwarzstein M, Tsugita A, Zabeau M (1984) Characterization of the genes coding for the EcoRV restriction and modification system of Escherichia coli. Gene 12:3659–3676
Brenner V, Venetianer P, Kiss A (1990) Cloning and nucleotide sequence of the gene encoding the EcaI DNA methyltransferase. Nucleic Acids Res 18:355–359
Brooks JE, Blumenthal RM, Gingeras TR (1983) The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res 11:837–851
Brooks JE, Benner JS, Heiter DF, Silber KR, Sznyter LA, Jager-Quinton T, Moran LS, Slatko BE, Wilson GG, Nwankwo DO (1989) Cloning the BamHI restriction modification system 17:979–997
Buhk H-J, Behrens B, Tailor R, Wilke K, Prada JJ, Gunthert U, Noyer-Weidner M, Jentsch S, Trautner TA (1984) Restriction and modification in Bacillus subtilis: nucleotide sequence, functional organization and product of the DNA methyltransferase gene of bacteriophage SPR. Gene 29:51–61
Cantoni GL (1985) The role of S-adenosylhomocysteine in the biological utilization of S-adenosylmethionine. In: Cantoni GL Razin A (eds) Biochemistry and biology of DNA methylation. Liss, New York, pp 47–65
Card CO, Wilson GG, Weule K, Hasapes J, Kiss A, Roberts RJ (1990) Cloning and characterization of the HpaII methylase gene. Nucleic Acids Res 18:1377–1383
Caserta M, Zacharias W, Nwankwo D, Wilson GG, Wells RD (1987) Cloning, sequencing, in vivo promoter mapping, and expression in Escherichia coli of the gene for the HhaI methyltransferase. J Biol Chem 262:4770–4777
Chandrasegaran S, Smith HO (1988) Amino acid sequence homologies among twenty-five restriction endonucleases and methylases. In: Sarma RH, Sarma MH (eds) Structure and expression, vol I: From proteins to ribosomes. Adenine Press, pp 149–156
Chandrasegaran S, Lunnen KD, Smith HO, Wilson GG (1988a) Cloning and sequencing the HinII restriction and modification genes. Gene 70:387–392
Chandrasegaran S, Wu, LP, Valda E, Smith HO (1988b) Overproduction and purification of the M.HhaII methyltransferase from Haemophilus haemolyticus. Gene 74:15–21
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890
Ehrlich M, Wilson GG, Kuo KC, Gehrke CW (1987) N4-methylcytosine as a minor base in bacterial DNA. J Bacteriol 169:939–943
Guschlbauer W (1988) The DNA and S-adenosylmethionine-binding regions of EcoDam and related methyltransferases. Gene 74:211–214
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580
Hanek T, Gerwin N, Fritz H-J (1989) Nucleotide sequence of the dcm locus of Escherichia coli K12. Nucleic Acids Res 17:5844
Hattman S (1981) DNA methylation. In: Boyer H (ed) The enzymes, vol XIV. Academic Press, New York, pp 517–548
Heidmann S, Seifert W, Kessler C, Domdey H (1989) Cloning, characterization and heterologous expression of the SmaI restriction-modification system. Nucleic Acids Res 17:9783–9796
Howard KA, Card C, Benner JS, Callahan HL, Maumus R, Silber K, Wilson G, Brooks JE (1986) Cloning the DdeI restriction-modification system using a two-step method. Nucleic Acids Res 14:7939–7951
Kaszubska W, Aiken C, O'Connor CD, Gumport RI (1989) Purification, cloning and sequence analysis of RsrI DNA methyltransferase: lack of homology between two enzymes, RsrI and EcoRI, that methylate the same nucleotide in identical recognition sequences. Nucleic Acids Res 17:10403–10425
Kiss A, Posfai G, Keller CC, Venetianer P, Roberts RJ (1985) Nucleotide sequence of the BsuRI restriction-modification system. Nucleic Acids Res 13:6403–6421
Kita K, Kotani H, Sugisaki H, Takanami M (1989) The FokI restriction-modification system I. Organization and nucleotide sequences of the restriction and modification genes. J Biol Chem 264:5751–5756
Klimasauskas S, Timinskas A, Menkevicius S, Butkiene D, Butkus V, Janulaitis A (1989) Sequence motifs characteristic of DNA [cytosine-N4] methyltransferases:similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res 17:9823–9832
Koob M, Grimes E, Szybalski W (1988) Conferring operator specificity on restriction endonucleases. Science 241:1084–1086
Kupper D, Zhou J-G, Venetianer P, Kiss A (1989) Cloning and structure of the BepI modification methylase. Nucleic Acids Res 17:1077–1088
Lacks SA, Mannarelli BM, Springhorn SS, Greenberg B (1986) Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: An intercellular cassette mechanism. Cell 46:993–1000
Landry D, Looney MC, Feehery GR, Slatko BE, Jack WE, Schildkraut I, Wilson GG (1989) M.FokI methylates adenine in both strands of its asymmetric recognition sequence. Gene 77:1–10
Lau PCK, Condie JA (1989) Nucleotide sequences from the colicin E5, E6 and E9 operons: Presence of a degenerate transposon-like structure in the ColE9-J plasmid. Mol Gen Genet 217:269–277
Lauster R (1989) Evolution of type II DNA methyltransferases: A gene duplication model. J Mol Biol 206:313–321
Lauster R, Kriebardis A, Guschlbauer W (1987) The GATATC-modification enzyme EcoRV is closely related to the GATC-recognizing methyltransferases DpnII and dam from E. coli and phage T4. FEBS Lett 220:167–176
Lauster R, Trautner TA, Noyer-Weidner M (1989) Cytosine-specific type II DNA methyltransferases: A conserved enzyme core with variable target-recognizing domains. J Mol Biol 206:305–313
Lin PM, Lee CH, Roberts RJ (1989) Cloning and characterization of the genes encoding the MspI restriction modification system. Nucleic Acids Res 17:3001–3011
Looney MC, Moran LS, Jack WE, Feehery GR, Benner JS, Slatko BE, Wilson GG (1989) Nucleotide sequence of the FokI restriction-modification system: Separate strand-specificity domains in the methyltransferase. Gene 80:193–208
Lunnen KD, Barsomoniam JM, Camp RR, Card CO, Chen S-Z, Croft R, Looney MC, Meda MM, Moran LS, Nwankwo DO, Slatko BE, Van Cott EM, Wilson GG (1988) Cloning type II restriction and modification genes. Gene 74:25–32
Macdonald PM, Mosig G (1984) Regulation of a new bacteriophage T4 gene, 69, that spans an origin of DNA replication. EMBO J 3:2863–2871
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning A laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, New York
Mannarelli BM, Balganesh TS, Greenberg B, Springhorn SS, Lacks SA (1985) Nucleotide sequence of the DpnII DNA methylase gene of Streptococcus pneumoniae and its relationship to the dam gene of Escherichia coli. Proc Natl Acad Sci USA 82:4464–4472
Marinus MG (1987) Methylation of DNA. In: Neidhardt FC (ed) Escherichia coli and Salmonella typhimurium, Cellular and molecular biology, vol. 1. American Society for Microbiology, Washington DC, pp 697–702
Nelson M, McClelland M (1989) Effect of site-specific methylation on DNA modification methyltransferases and restriction endonucleases. Nucleic Acids Res (Suppl): 17:r389-r415
Posfai J, Bhagwat AS, Posfai G, Roberts RJ (1989) Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res 17:2421–2435
Qiang B-Q, Schildkraut I (1986) Two unique restriction endonucleases from Neisseria lactamica. Nucleic Acids Res 14:1991–1999
Raleigh EA, Wilson G (1986) Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci USA 83:9070–9074
Roberts RJ (1989) Restriction enzymes and their isoschizomers. Nucleic Acids Res (Suppl) 17: r247-r387
Smith HO, Annau TM, Chandrasegaran S (1990) Finding sequence motifs in groups of functionally related proteins. Proc Natl Acad Sci USA 87:826–830
Som S, Bhagwat AS, Friedman S (1987) Nucleotide sequence and expression of the gene encoding the EcoRII modification enzyme. Nucleic Acids Res 15:313–332
Sugisaki H, Kita K, Takanami M (1989) The FokI restriction-modification system II. Presence of two domains in FokI methylase responsible for modification of different DNA strands. J Biol Chem 264:5757–5761
Tao T, Walter J, Brennan KJ, Cotterman MM, Blumenthal RM (1989) Sequence, internal homology and high-level expression of the gene for a DNA-(cytosine N4)-methyltransferase, M.PvuII. Nucleic Acids Res 17:4161–4175
Trautner TA, Balganesh TS, Pawlek B (1988) Chimeric multispecific DNA methyltransferases with novel combinations of target recognition. Nucleic Acids Res 16:6649–6658
Vieira J, Messing J (1982) The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259–268
Walter J, Noyer-Weider M, Trautner TA (1990) The amino acid sequence of the CCGG recognizing DNA methyltransferase M.BsuFI: implications for the analysis of sequence recognition by cytosine DNA methyltransferases. EMBO J 9:1007–1013
Weil MD, McClelland M (1989) Enzymatic cleavage of a bacterial genome at a 10-base-pair recognition site. Proc Natl Acad Sci USA86:51–55
Wilke K, Rauhut E, Noyer-Weidner M, Lauster R, Pawlek B, Behrens B, Trautner TA (1988) Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J 7:2601–2609
Wilson GG (1988a) Type II restriction-modification system. Trends Genet 4:314–318
Wilson GG (1988b) Cloned restriction-modification systems — a review. Gene 74:281–289
Yanisch-Perron C, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the Ml3mp18 and pUC19 vectors. Gene 33:103–119
Author information
Authors and Affiliations
Additional information
Communicated by W. Goebel
Issued as NRCC publication number 31454
Rights and permissions
About this article
Cite this article
Labbé, D., Höltke, H.J. & Lau, P.C.K. Cloning and characterization of two tandemly arranged DNA methyltransferase genes of Neisseria lactamica: An adenine-specific M.NlaIII and a cytosine-type methylase. Molec. Gen. Genet. 224, 101–110 (1990). https://doi.org/10.1007/BF00259456
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00259456