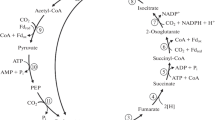
Abstract
The extremely thermophilic, obligately aerobic bacterium Sulfolobus solfataricus forms the tetrapyrrole precursor, δ-aminolevulinic acid (ALA), from glutamate by the tRNA-dependent five-carbon pathway. This pathway has been previously shown to occur in plants, algae, and most prokaryotes with the exception of the α-group of proteobacteria (purple bacteria). An alternative mode of ALA formation by condensation of glycine and succinyl-CoA occurs in animals, yeasts, fungi, and the α-proteobacteria. Sulfolobus and several other thermophilic, sulfur-dependent bacteria, have been variously placed within a subgroup of archaea (archaebacteria) named crenarchaeotes, or have been proposed to comprise a distinct prokaryotic group designated eocytes. On the basis of ribosomal structure and certain other criteria, eocytes have been proposed as predecessors of the nuclear-cytoplasmic descent line of eukaryotes. Because aplastidic eukaryotes differ from most prokaryotes in their mode of ALA formation, and in view of the proposed affiliation of eocytes to eukaryotes, it was of interest to determine how eocytes form ALA. Sulfolobus extracts were able to incorporate label from [1-14C]glutamate, but not from [2-14C]glycine, into ALA. Glutamate incorporation was abolished by preincubation of the extract with RNase. Sulfolobus extracts contained glutamate-1-semialdehyde aminotransferase activity, which is indicative of the five-carbon pathway. Growth of Sulfolobus was inhibited by gabaculine, a mechanism-based inhibitor of glutamate-1-semialdehyde aminotransferase, an enzyme of the five-carbon ALA biosynthetic pathway. These results indicate that Sulfolobus uses the five-carbon pathway for ALA formation.
Similar content being viewed by others
Abbreviations
- AHA:
-
4-amino-5-hexynoic acid
- ALA:
-
δ-aminolevulinic acid, Gabaculine, 3-amino-2,3-dihydrobenzoic acid
- GSA:
-
glutamate 1-semialdehyde
References
Anemüller S, Schäfer G (1990) Cytochrome aa 3 from Sulfolobus acidocaldarius. A single-subunit, quinol-oxidizing archaebacterial terminal oxidase. Eur J Biochem 191: 297–305.
Anemüller S, Lübben M, Schäfer G (1985) The respiratory system of Sulfolobus acidocaldarius, a thermoacidophilic archaebacterium. FEBS Lett 193: 83–87.
Avissar YJ (1980) Biosynthesis of 5-aminolevulinate from glutamate in Anabaena variabilis. Biochim Biophys Acta 613: 220–228.
Avissar YJ, Beale SI (1989a) The aminotransferase step in the formation of δ-aminolevulinic acid from glutamate: isolation of the enzyme from Chlorella vulgaris, requirement for pyridoxal phosphate, and inhibition by gabaculine and acetylenic GABA. Plant Physiol 89: S51.
Avissar YJ, Beale SI (1989b) Biosynthesis of tetrapyrrole pigment precursors: pyridoxal requirement of the aminotransferase step in the formation of δ-aminolevulinate from glutamate in extracts of Chlorella vulgaris. Plant Physiol 89: 852–859.
Avissar YJ, Ormerod JG, Beale SI (1989) Distribution of δ-aminolevulinic acid biosynthetic pathways among photographic bacterial groups. Arch Microbiol 151: 513–519.
Beale SI (1990) Biosynthesis of the tetrapyrrole pigment precursor, δ-aminolevulinic acid, from glutamate. Plant Physiol 93: 1273–1279.
Beale SI, Castelfranco PA (1974) The biosynthesis of δ-aminolevulinic acid in higher plants. II. Formation of 14C-δ-aminolevulinic acid from labeled precursors in greening plant tissues. Plant Physiol 53: 297–303.
Beale SI, Foley T, Dzelzkalns V (1981) δ-Aminolevulinic acid synthase from Euglena gracilis. Proc Natl Acad Sci USA 78: 1666–1669.
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254.
Brock TD, Brock KM, Belly RT, Weiss RL (1972) Sulfolobus: a new genus of sulfuroxidizing bacteria living at low pH and high temperature. Arch Mikrobiol 84: 54–68.
Corriveau JL, Beale SI (1986) Influence of gabaculine on growth, chlorophyll synthesis, and δ-aminolevulinic acid synthase activity in Euglena gracilis. Plant Sci 45: 9–17.
Elich TD, Lagarias JC (1988) 4-Amino-5-hexynoic acid — a potent inhibitor of tetrapyrrole biosynthesis in plants. Plant Physiol 88: 747–751.
Elliott T, Avissar YJ, Rhie G, Beale SI (1990) Cloning and sequence of the Salmonella typhimurium hemL gene and identification of the missing enzyme in hemL mutants as glutamate-1-semialdehyde aminotransferase. J Bacteriol 172: 7071–7084.
Fox GE, Stackebrandt E, Hespell RB, Gibson J, Maniloff J, Dyer TA, Wolfe RS, Balch WE, Tanner RS, Magrum LJ, Zablen LB, Blakemore R, Gupta R, Bonen L, Lewis BJ, Stahl DA, Luehrsen KR, Chen KN, Woese CR (1980) The phylogeny of prokaryotes. Science 209: 457–463.
Friedmann HC, Thauer RK (1986) Ribonuclease-sensitive δ-aminolevulinic acid formation from glutamate in cell extracts of Methanobacterium thermoautotrophicum. FEBS Lett 207: 84–88.
Friedmann HC, Thauer RK, Gough SP, Kannangara CG (1987) δ-Aminolevulinic acid formation in the archaebacterium Methanobacterium thermoautotrophicum requires tRNAGlu. Carlsberg Res Commun 52: 363–371.
Gibson KD, Laver WG, Neuberger A (1958) Initial stages in the biosynthesis of porphyrins. 2 The formation of δ-aminolaevulic acid from glycine and succinyl-coenzyme A by particles from chicken erythrocytes. Biochem J 70: 71–81.
Gough SP, Kannangara CG, Bock K (1989) A new method for the synthesis of glutamate 1-semialdehyde. Characterization of its structure in solution by NMR spectroscopy. Carlsberg Res Commun 54: 99–108.
Guikema JA, Freeman L, Fleming EH (1986) Effects of gabaculine on pigment biosynthesis in normal and nutrient deficient cells of Anacystis nidulans. Plant Physiol 82: 280–284.
Hoober JK, Kahn A, Ash D, Gough S, Kannangara CG (1988) Biosynthesis of δ-aminolevulinate in greening barley leaves. IX. Structure of the substrate, mode of gabaculine inhibition, and the catalytic mechanism of glutamate 1-semialdehyde amino-transferase. Carlsberg Res Commun 53: 11–25.
Hoult RC, Rees D, Rogers LJ, Smith AJ (1986) Specific inhibition of tetrapyrrole biosynthesis by 3-amino 2,3-dihydrobenzoic acid (gabaculin) in the cyanobacterium Synechococcus 6301. Arch Microbiol 146: 57–62.
Kannangara CG, Schouboe A (1985) Biosynthesis of δ-aminolevulinate in greening barley leaves. VII. Glutamate 1-semialdehyde accumulation in gabaculine treated leaves. Carlsberg Res Commun 50: 179–191.
Kikuchi G, Kumar A, Talmage P, Shemin D (1958) The enzymatic synthesis of δ-aminolevulinic acid. J Biol Chem 233: 1214–1249.
Lake JA (1988) Origin of eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature 331: 184–186.
Lake JA, Henderson E, Oakes M, Clark MW (1984) Ecocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci USA 81: 3786–3790.
Li J-M, Brathwaite O, Cosloy SD, Russell CS (1989) 5-Aminolevulinic acid synthesis in Escherichia coli. J Bacteriol 171: 2547–2552.
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
Mayer SM, Weinstein JD, Beale SI (1987) Enzymatic conversion of glutamate to δ-aminolevulinate in soluble extracts of Euglena gracilis. J Biol Chem 262: 12541–12549.
Meller E, Harel E (1978) The pathway of 5-aminolevulinic acid synthesis in Chlorella vulgaris and in Fremyella diplosiphon. In: Akoyunoglou G, Argyroudi-Akoyunoglou JH (eds) Chloroplast development. Elsevier, Amsterdam, pp 51–57.
Murakami K, Hashimoto Y, Murooka Y (1993a) Cloning and characterization of the gene encoding glutamate 1-semialdehyde 2,1-aminomutase, which is involved in δ-aminolevulinic acid synthesis in Propionibacterium freudenreichii. Appl Environ Microbiol 59: 347–350.
Murakami K, Korbsrisate S, Asahara N, Hashimoto Y, Murooka Y (1993b) Cloning and characterization of the glutamate 1-semialdehyde aminomutase gene from Xanthomonas campestris pv. phaseoli. Appl Microbiol Biotechnol 38: 502–506.
Oh-hama T, Seto H, Miyachi S (1986a) 13C-NMR evidence for bacteriochlorophyll c formation by the C5 pathway in green sulfur bacterium, Prosthecochloris. Eur J Biochem 159: 189–194.
Oh-hama T, Seto H, Miyachi S (1986b) 13C-NMR evidence of bacteriochlorophyll a formation by the C5 pathway in Chromatium. Arch Biochem Biophys 246: 192–109.
Oh-hama T, Stolowich NJ, Scott AI (1988) 5-Aminolevulinic acid formation from glutamate via the C5 pathway in Clostridium thermoaceticum. FEBS Lett 228: 89–93.
O'Neill GP, Chen M-W, Söll D (1989) δ-Aminolevulinic acid biosynthesis in Escherichia coli and Bacillus subtilis involves formation of glutamyl-tRNA. FEMS Microbiol Lett 60: 255–260.
Pace NR, Olsen GJ, Woese CR (1986) Ribosomal RNA phylogeny and the lines of evolutionary descent. Cell 45: 325–326.
Redlinger RE, Beale SI (1991) Evolution of chlorophyll structure and function. In: Holmes MG (ed) Photoreceptor evolution and function. Academic Press, New York, pp 151–179.
Rivera MC, Lake JA (1992) Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 257: 74–76.
Smith KM, Huster MS (1987) Bacteriochlorophyll-c formation via the glutamate C-5 pathway in Chlorobium bacteria. J Chem Soc Chem Commun 1987: 14–16.
Walsh CT (1984) Suicide substrates, mechanism-based enzyme inactivators: recent developments. Annu Rev Biochem 53: 493–535.
Weinstein J, Beale SI (1983) Separate physiological roles and subcellular compartments for two tetrapyrrole biosynthetic pathways in Euglena gracilis. J Biol Chem 258: 6799–6807.
Weinstein JD, Beale SI (1985a) Enzymatic conversion of glutamate to δ-aminolevulinate in soluble extracts of the unicellular green alga, Chlorella vulgaris. Arch Biochem Biophys 237: 454–464.
Weinstein JD, Beale SI (1985b) RNA is required for enzymic conversion of glutamate to δ-aminolevulinic acid by extracts of Chlorella vulgaris. Arch Biochem Biophys 239: 87–93.
Woese CR (1987) Bacterial evolution. Microbiol Rev 51: 221–271.
Woese CR, Kandler O, Wheelis ML (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad Sci USA 87: 4576–4579.
Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR (1985) Mitochondrial origins. Proc Natl Acad Sci USA 82: 4443–4447.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Matters, G.L., Beale, S.I. Biosynthesis of δ-aminolevulinic acid from glutamate by Sulfolobus solfataricus . Arch. Microbiol. 161, 272–276 (1994). https://doi.org/10.1007/BF00248704
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00248704