Abstract
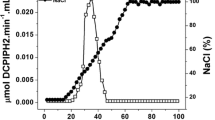
The localization of hydrogenase protein in Desulfovibrio gigas cells grown either in lactate-sulfate or hydrogen-sulfate media, has been investigated by subcellular fractionation with immunoblotting and by electron microscopic immunocytochemistry. Subcellular fractionation experiments suggest that no integral membrane-bound hydrogenase is present in D. gigas. About 40% of the hydrogenase activity could be extracted by treatment of D. gigas cells with Tris-EDTA buffer. The rest of the soluble hydrogenase activity (50%) was found in the soluble fraction which was obtained after disruption of Tris-EDTA extracted cells and high speed centrifugation. Both soluble hydrogenase fractions purified to homogeneity showed identical molecular properties including the N-terminal aminoacid sequences of their large and small subunits. Polyacrylamide gel electrophoresis of the proteins of the subcellular fractions revealed a single band of hydrogenase activity exhibiting the same mobility as purified D. gigas hydrogenase. Western blotting carried out on these subcellular fractions revealed crossreactivity with the antibodies raised against (NiFe) hydrogenase. The lack of crossreactivity with antibodies against (FE) or (NiFeSe) hydrogenases, indicated that only (NiFe) type hydrogenase is present in D. gigas.
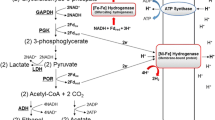
Immunocytolocalization in ultrathin frozen sections of D. gigas cells grown either in lactate-sulfate, pyruvate-sulfate or hydrogen-sulfate media showed only a (NiFe) hydrogenase located in the periplasmic space. The bioenergetics of D. gigas are discussed in the light of these findings.
Similar content being viewed by others
References
Adams MWW, Hall DO (1979) Properties of the solubilized membrane-bound hydrogenase from the photosynthetic bacterium Rhodospirillum rubrum. Arch Biochem Biophys 195:288–299
Anba J, Bernadac A, Pages JM, Lazdunski C (1984) The periseptal annulus in Escherichia coli. Biol Cell 50:273–278
Badziong W, Thauer RK (1980) Vectorial electron transport in Desulfovibrio vulgaris (Marburg) growing on hydrogen plus sulfate as sole energy source. Arch Microbiol 125:167–174
Bell GR, LeGall J, Peck HD (1974) Evidence for the periplasmic location of hydrogenase in Desulfovibrio gigas. J Bacteriol 120:994–997
Brandis A, Thauer RK (1981) Growth of Desulfovibrio species on hydrogen and sulfate as sole energy source. J Gen Microbiol 126:249–252
Bryant MP, Campbell LL, Reddy CA, Crabill MR (1977) Growth of Desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria. Appl Environ Microbiol 33:1162–1169
Cammack R, Patil D, Aguirre R, Hatchikian EC (1982) Redox properties of the ESR-detectable nickel in hydrogenase from Desulfovibrio gigas. FEBS Lett 142:289–292
Cypionka H, Dilling W (1986) Intracellular localization of the hydrogenase in Desulfotomaculum orientis. FEMS Lett 36:257–260
Czechowski MH, He SH, Nacro M, DerVartanian DV, Peck HD, LeGall J (1984) A cytoplasmic Ni−Fe hydrogenase with high specific activity from Desulfovibrio multispirans sp.n., a new species of sulfate-reducing bacterium. Biochem Biophys Res Commun 125:1024–1032
Davis BJ (1964) Disc electrophoresis. II. Method and application to human serum proteins. Ann NY Acad Sci 121:404–427
Fauque G, Peck HD, Moura JJG, Huynh BH, Berlier Y, DerVartanian DV, Teixeira M, Przybyla AE, Lespinat PA, Moura I, LeGall J (1988) The three classes of hydrogenases from sulfate-reducing bacteria of the genus Desulfovibrio. FEMS Microbiol Rev 54:299–344
Fernandez VM, Hatchikian C, Cammack R (1985) Properties and reactivation of two different deactivated forms of Desulfovibrio gigas hydrogenase. Biochim Biophys Acta 832:69–70
Fernandez VM, Rua ML, Reyes P, Cammack R, Hatchikian EC (1989) Inhibition of Desulfovibrio gigas hydrogenase with copper salts and other metal ions. Eur J Biochem 185:449–454
Glick BR, Martin WG, Martin SM (1980) Purification and properties of the periplasmic hydrogenase from Desulfovibrio desulfuricans. Can J Microbiol 26:1214–1223
Hatchikian EC, Bruschi M, LeGall J (1978) Characterization of the periplasmic hydrogenase from Desulfovibrio gigas. Biochem Biophys Res Commun 82:451–461
Hatchikian EC, Fernandez VM, Cammack R (1990a) The hydrogenases of sulfate-reducing bacteria: physiological, biochemical and catalytic aspects. In: Belaich JP, Brushi M, Garcia JL (eds) Microbiology and biochemistry of strict anaerobes involved in interspecies H2-transfer. FEMS Symposium. Plenum Press, New York, pp 53–73
Hatchikian EC, Traore A, Fernandez VM, Cammack R (1990b) Characterization of the nickel-iron periplasmic hydrogenase from Desulfovibrio fructosovorans. Eur J Biochem 187:635–643
Hawkes R, Niday E, Gordon J (1982) A dot-immunobinding assay for monoclonal and other antibodies. Anal Biochem 119:142–147
Kremer DR, Veenhuis M, Fauque G, Peck HD, LeGall J, Lampreia J, Moura JJG, Hansen TA (1988) Immunocytochemical localization of APS reductase and bisulfite reductase in three Desulfovibrio species. Arch Microbiol 150:296–301
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature 227:680–685
Lalla-Maharaih WV, Hall DO, Cammack R, Rao KK, LeGall J (1983) Purification and properties of the membrane bound hydrogenase from Desulfovibrio desulfuricans. Biochem J 209:445–454
Lee JP, Peck HD (1971) Purification of the enzyme reducing bisulfite to trithionate from Desulfovibrio gigas and its identification as desulfoviridin. Biochem Biophys Chem Commun 45:583–589
LeGall J, Mazza G, Dragoni N (1965) Le cytochrome c3 de Desulfovibrio gigas. Biochim Biophys Acta 99:385–387
Lissolo T, Choi ES, LeGall J, Peck HD (1986) The presence of multiple intrinsic membrane nickel-containing hydrogenases in Desulfovibrio vulgaris (Hildenborough). Biochem Biophys Res Commun 139:701–708
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Lupton FS, Conrad R, Zeikus JG (1984) Physiological function of hydrogen metabolism during growth of sulfidogenic bacteria on organic substrates. J Bacteriol 159:843–849
Magee EL, Ensley BD, Barton LL (1978) An assessment of growth yields and energy coupling in Desulfovibrio. Arch Microbiol 117:21–26
Moura JJG, Moura I, Huynh BH, Krüger HJ, Teixeira M, DuVarney RC, DerVartanian DV, Xavier AV, Peck HD, LeGall J (1982) Unambiguous identification of the nickel EPR signal in 61Ni-enriched Desulfovibrio gigas hydrogenase. Biochem Biophys Res Commun 108: 1388–1393
Nivière V, Forget N, Bovier-Lapierre G, Bonicel J, Hatchikian EC (1988) Isolation, aminoacid analysis and N-terminal sequence determination of the two subunits of the mickel-containing hydrogenase of Desulfovibrio gigas. Biochimie 70:267–271
Nivière V, Hatchikian EC, Bianco P, Haladjian F (1988) Kinetic studies of electron transfer between hydrogenase and cytochrome c3 from Desulfovibrio gigas. Electrochemical properties of cytochrome c3. Biochim Biophys Acta 935:34–40
Odom JM, Peck HD (1981a) Hydrogen cycling as a general mechanism for energy coupling in the sulfate-reducing bacteria Desulfovibrio sp. FEMS Microbiol Lett 12:47–50
Odom JH, Peck HD (1981b) Localization of dehydrogenases, reductases, and electron transfer components in the sulfate-reducing bacterium Desulfovibrio gigas. J Bacteriol 147:161–169
Odom JM, Peck HD (1984) Hydrogenase, electron-transfer proteins and energy coupling in the sulfate-reducing bacteria Desulfovibrio. Ann Rev Microbiol 38:551–592
Peck HD, LeGall J, Lespinat PA, Berlier Y, Fauque G (1987) A direct demonstration of hydrogen cycling by Desulfovibrio vulgaris employing membrane-inlet mass spectrometry. FEMS Microbiol Letters 40:295–299
Prickril BC, He S-H, Li C, Menon N, Choi E-S, Przybyla AE, DerVartanian DV, Peck HD, Fauque G, LeGall J, Teixeira M, Moura I, Moura JJG, Patil D, Huynh B (1987) Identification of three classes of hydrogenase in the genus Desulfovibrio. Biochem Biophys Res Commun 149:369–377
Rieder R, Cammack R, Hall DO (1984) Purification and properties of the soluble hydrogenase from Desulfovibrio desulfuricans Norway 4. Eur J Biochem 145:637–643.
Rohde M, Fürstenau U, Mayer F, Przybyla AE, Peck HD, LeGall J, Choi ES, Menon NK (1990) Localization of membrane-associated (NiFe) and (NiFeSe) hydrogenases of Desulfovibrio vulgaris using immunoelectron microscopic procedures. Eur J Biochem 191:389–396
Teixeira M, Fauque G, Moura I, Lespinat PA, Berlier Y, Prickril B, Peck HD, Xavier AV, LeGall J, Moura JJG (1987) Nickel-(ironsulfur)-selenium containing hydrogenases from Desulfovibrio baculatus (DSM 1743). Eur J Biochem 167:47–58
Towbin H, Staehlin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76:4350–4354
Traore AS, Hatchikian EC, Belaich JP, LeGall J (1981) Microcalorimetric studies of the growth of sulfate-reducing bacteria: energetics of Desulfovibrio vulgaris growth. J Bacteriol 145:191–199
Traore SA, Fardeau ML, Hatchikian EC, LeGall J, Bélaich JP (1983) Energetics of growth of a defined mixed culture of Desulfovibrio vulgaris and Methanosarcina barkeri: interspecies hydrogen transfer in batch and continuous cultures. Appl Environ Microbiol 46:1152–1156
Van derWesten H, Mayhew SG, Veeger C (1978) Separation of hydrogenase from intact cells of Desulfovibrio vulgaris. Purification and properties. FEBS Lett 86:122–126
Voordouw G, Kent HM, Postgate JR (1987) Identification of the genes for hydrogenase and cytochrome c3 in Desulfovibrio. Can J Microbiol 33:1006–1010
Voordouw G, Nivière V, Ferris FG, Fedorak PM, Westlake DWS (1990) Distribution of hydrogenase genes in Desulfovibrio spp. and their use in identification of species from the oil field environment. Appl Environ Microbiol 56:3748–3754
Zeikus JG, Fuchs G, Kenealy W, Thauer RK (1977) Oxidoreductases involved in cell carbon synthesis of Methanobacterium thermoautotrophicum. J Bacteriol 132:604–613
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Nivière, V., Bernadac, A., Forget, N. et al. Localization of hydrogenase in Desulfovibrio gigas cells. Arch. Microbiol. 155, 579–586 (1991). https://doi.org/10.1007/BF00245353
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00245353