Abstract
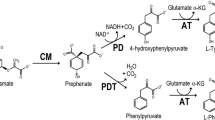
Extensive diversity in features of aromatic amino acid biosynthesis and regulation has become recognized in eubacteria, but almost nothing is known about the extent to which such diversity exists within the archaebacteria. Methanohalophilus mahii, a methylotrophic halophilic methanogen, was found to synthesize l-phenylalanine and l-tyrosine via phenylpyruvate and 4-hydroxyphenylpyruvate, respectively. Enzymes capable of using l-arogenate as substrate were not found. Prephenate dehydrogenase was highly sensitive to feedback inhibition by l-tyrosine and could utilize either NADP+ (preferred) or NAD+ as cosubstrate. Tyrosine-pathway dehydrogenases having the combination of narrow specificity for a cyclohexadienyl substrate but broad specificity for pyridine nucleotide cofactor have not been described before. The chorismate mutase enzyme found is a member of a class which is insensitive to allosteric control. The most noteworthy character state was prephenate dehydratase which proved to be subject to multimetabolite control by feedback inhibitor (l-phenylalanine) and allosteric activators (l-tyrosine, l-tryptophan, l-leucine, l-methionine and l-isoleucine). This interlock type of prephenate dehydratase, also known to be broadly distributed among the gram-positive lineage of the eubacteria, was previously shown to exist in the extreme halophile, Halobacterium vallismortis. The results are consistent with the conclusion based upon 16S rRNA analyses that Methanomicrobiales and the extreme halophiles cluster together.
Similar content being viewed by others
Abbreviations
- DAHP:
-
3-deoxy-d-arabino-heptulosonate-7-phosphate
References
Ahmad S, Jensen RA (1988) New prospects for deducing the evolutionary history of metabolic pathways in prokaryotes: aromatic biosynthesis as a case-in-point. Orig Life Evol Biosphere 18: 41–57.
Ahmad S, Weisburg WG, Jensen RA (1990) Evolution of aromatic amino acid biosynthesis and application to the fine-tuned phylogenetic positioning of enteric bacteria. J Bacteriol 172: 1051–1061.
Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS (1979) Methanogenesis: reevaluation of a unique biological group. Microbiol Rev 43: 260–296.
Berry A, Ahmad S, Liss A, Jensen RA (1987) Enzymological features of aromatic amino acid biosynthesis reflect the phylogeny of mycoplasmas. J Gen Microbiol 133: 2147–2154.
Bonner CA, Jensen RA (1987) Arogenate dehydrogenase. Methods Enzymol 142: 488–494.
Bonner CA, Fischer RS, Ahmad S, Jensen RA (1990) Remnants of an ancient pathway to l-phenylalanine and l-tyrosine in enteric bacteria: evolutionary implications and biotechnological impact. Appl Environ Microbiol 56: 3741–3747.
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254.
Byng GS, Kane JF, Jensen RA (1982) Diversity in the routing and regulation of complex biochemical pathways as indicators of microbial relatedness. CRC Crit Rev Microbiol 9: 227–252.
Champney WS, Jensen RA (1970) The enzymology of prephenate dehydrogenase in Bacillus subtilis. J Biol Chem 245: 3763–3770.
Cotton RGH, Gibson F (1965) The biosynthesis of phenylalanine and tyrosine: enzymes converting chorismic acid into prephenic acid and their relationships to prephenate dehydratase and prephenate dehydrogenase. Biochim Biophys Acta 100: 76–88.
Danson MJ (1988) Archaebacteria: the comparative enzymology of their central metabolic pathways. Adv Microbiol Physiol 29: 165–231.
Fazel AM, Jensen RA (1980) Regulation of prephenate dehydratase in coryneform species of bacteria by l-phenylalanine and by remote effectors. Arch Biochem Biophys 200: 165–176.
Fazel AM, Bowen JR, Jensen RA (1980) Arogenate (pretyrosine) is an obligatory intermediate of l-tyrosine biosynthesis: confirmation in a microbial mutant. Proc Natl Acad Sci USA 77: 1270–1273.
Fischer RS, Jensen RA (1987a) Arogenate dehydratase. Methods Enzymol 142: 495–502.
Fischer RS, Jensen RA (1987b) Prephenate dehydrogenase (monofunctional). Methods Enzymol 142: 503–507.
Fischer RS, Jensen RA (1987c) Prephenate dehydratase (monofunctional). Methods Enzymol 142: 507–512.
Gibson F (1964) Chorismic acid: purification and some chemical and physical studies. Biochem J 90: 256–257.
Hall GC, Jensen RA (1980) Enzymological basis for growth inhibition by l-phenylalanine in the cyanobacterium Synechocystis sp. 29108. J Bacteriol 144: 1034–1042.
Hall GC, Flick MB, Gherna RL, Jensen RA (1982) Biochemical diversity for biosynthesis of aromatic amino acids among the cyanobacteria. J Bacteriol 149: 65–78.
Jensen RA (1969) Metabolic interlock: regulatory interactions exerted between biochemical pathways. J Biol Chem 244: 2816–2823.
Jensen RA (1976) Enzyme recruitment in evolution of new function. Annu Rev Microbiol 30: 409–425.
Jensen RA (1985) Biochemical pathways can be traced backward through evolutionary time. Mol Biol Evol 2: 92–108.
Jensen RA (1992) An emerging outline of the evolutionary history of aromatic amino acid biosynthesis. In: Mortlock RP (ed) The evolution of metabolic function. CRC Press, Boca Raton, Florida, pp 205–236.
Jensen RA, Nester EW (1966) Regulatory enzymes of aromatic amino acid biosynthesis in Bacillus subtilis. I. Purification and properties of DAHP synthetase. J Biol Chem 241: 3365–3372.
Jensen RA, d'Amato TA, Hochstein LI (1988) An extreme-halophile archaebacterium possesses the interlock type of prephenate dehydratase characteristic of the gram-positive eubacteria. Arch Microbiol 148: 365–371.
Lai M, Sowers KR, Robertson DE, Roberts MF, Gunsalus RP (1991) Distribution of compatible solutes in the halophilic methanogenic archaebacteria. J Bacteriol 173: 5352–5358.
Lake JA, Clark MW, Henderson E, Fay SP, Oakes M, Scheinman A, Thornber JP, Mah RA (1985) Eubacteria, halobacteria, and the origin of photosynthesis: the photocytes. Proc Natl Acad Sci USA 82: 3716–3720.
Paterek JR, Smith PH (1985) Isolation and characterization of a halophilic methanogen from great salt lake. Appl Env Microbiol 50: 877–881.
Paterek JR, Smith PH (1985) Methanohalophilus mahii gen. nov., sp. nov., a methylotrophic halophilic methanogen. Int J Syst Bacteriol 38: 122–123.
Petzel JP, Hartman PA (1991) Aromatic amino acid biosynthesis and carbohydrate catabolism in strictly anaerobic mollicutes (Anaeroplasma spp.). Syst Appl Microbiol 13: 240–247.
Pierson DL, Jensen RA (1974) Metabolic interlock: control of an interconvertible prephenate dehydratase by hydrophobic amino acids in Bacillus subtilis. J Mol Biol 90: 563–579.
Rebello JL, Jensen RA (1970) Metabolic interlock: the multimetabolite control of prephenate dehydratase activity in Bacillus subtilis. J Biol Chem 245: 3738–3744.
Riepl RG, Glover GI (1979) Regulation and state of aggregation of Bacillus subtilis prephenate dehydratase in the presence of allosteric effectors. J Biol Chem 254: 10321–10328.
White BN, Bayley STG (1972) Further codon assignments in an extremely halophilic bacterium using a cell-free protein-synthesizing system and a ribosomal binding assay. Can J Biochem 50: 600–609.
Woese CR (1987) Bacterial evolution. Microbiol Rev 51: 221–271.
Woese CR, Olsen GJ (1986) Archaebacterial phylogeny: perspectives on the urkingdoms. Syst Appl Microbiol 7: 163–177.
Yang DC, Kaine B, Woese CR (1985) The phylogeny of archaebacteria. Syst Appl Microbiol 6: 251–256.
Ycas M (1974) On earlier states of the biochemical system. J Theor Biol 44: 145–160.
Zaccai C, Eisenberg H (1990) Halophilic proteins and the influence of solvent on protein stabilization. Trends Biochem Sci 15: 333–337.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Fischer, R.S., Bonner, C.A., Boone, D.R. et al. Clues from a halophilic methanogen about aromatic amino acid biosynthesis in archaebacteria. Arch. Microbiol. 160, 440–446 (1993). https://doi.org/10.1007/BF00245304
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00245304