Abstract
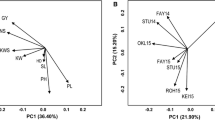
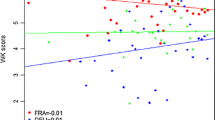
The additive main effects and multiplicative interaction (AMMI) model has emerged as a powerful analytical tool for genotype x environment studies. The objective of the present study was to assess its value in quantitative trait locus (QTL) mapping. This was done through the analysis of a large two-way table of genotype-by-environment data of barley (Hordeum vulgare L.) grain yields, where the genotypes constituted a genetic population suitable for mapping studies. Grain yield data of 150 doubled haploid lines derived from the ‘Steptoe’ x ‘Morex’ cross, and the two parental lines, were taken by the North American Barley Genome Mapping Project (NABGMP) at 16 environments throughout the barley production areas of the USA and Canada. Four regions of the genome were responsible for most of the differential genotypic expression across environments. They accounted for approximately 50% of the genotypic main effect and 30% of the genotype x environment interaction (GE) sums of squares. The magnitude and sign of AMMI scores for genotypes and sites facilitate inferences about specific interactions. The parallel use of classification (cluster analysis of environments) and ordination (principal component analysis of GE matrix) techniques allowed most of the variation present in the genotype x environment matrix to be summarized in just a few dimensions, specifically four QTLs showing differential adaptation to four clusters of environments. Thus, AMMI genotypic scores, when the genotypes constituted a population suitable for QTL mapping, could provide an adequate way of resolving the magnitude and nature of QTL x environment interactions.
Similar content being viewed by others
References
Chen F, Hayes PM (1989) A comparison of Hordeum bulbosum — mediated haploid production efficiency in barley using in vitro floret and tiller culture. Theor Appl Genet 77: 701–704
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971
Fox PN, Roseille AA (1982) Reducing the influence of environmental main effects in pattern analysis of plant breeding environments. Euphytica 31: 645–656
Fox PN, Crossa J, Romagosa I (1996) Multi-environment testing and genotype x environment interaction. In: Kempton RA, Fox PN (eds) Statistical methods for plant variety evaluation. Chapman and Hall. London (in press)
Gauch HG (1992) Statistical analysis of regional yield trials: AMMI analysis of factorial designs. Elsevier, Amsterdam
Haley CS, Knott SA (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324
Hayes PM, Iyambo O, the NABGMP (1993) Summary of QTL effects in the Steptoe x Morex population. Barley Genet Newslett 23: 98–143
Hayes PM, Liu BH, Knapp SJ, Chen FQ, Jones B, Blake TK, Franckowiak JD, Rasmusson DC, Sorrells M, Ullrich SE, Wesenberg D, Kleinhofs A (1993b) Quantitative trait locus effects and environmental interaction in a sample of North American barley germ plasm. Theor Appl Genet 87: 329–401
Hayes PM, Matthews D, The NABGMP (1994) Online datasets for the Steptoe x Morex barley mapping population. Files available via Gopher, host greengenes.cit.cornell.edu
Kleinhofs A (1995) Online datasets for the Steptoe x Morex barley map. File available via Gopher, host greengenes.cit.cornell.edu
Kleinhofs A, Kilian A, Saghai Maroof MA, Bitashev RM, Hayes PM, Chen FQ, Lapitan N, Fenwick A, Blake TK, Kanazin V, Ananiev E, Dahleen E, Kudrna D, Bollinger J, Knapp SJ, Liu B, Sorrells M, Heun M, Franckowiak JD, Hoffman D, Skadsen R, Steffenson RJ (1993) A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor Appl Genet 86: 705–712
Lander E, Green P, Abrahamson J, Barlow A, Daley M, Lincoln S, Newburg L (1987) MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181
Lincoln S, Daly M, Lander E (1992) Mapping genes controlling quantitative traits with MAPMAKER/QTL 1.1. Whitehead Institute Technical Report. 2nd edn
Mather DE (1995) Online data set for the Steptoe/Morex base map. File available via ftp.gnome.agrenv.mcgill.ca
Ozdemir E (1994) Genetic analysis of malt extract and grain yield in a spring barley (Hordeum vulgare L.) cross. MS thesis, Oregon State University
Romagosa I, Fox PN (1993) Genotye x environment interaction and adaptation. In: Hayward MD, Bosemark NO, Romagosa I (eds) Plant breeding: principles and prospects. Chapman and Hall, London, pp 373–390
Royo C, Rodriguez A, Romagosa I (1993) Adaptation of complete and substituted triticale to acid soils. Plant Breed 111: 113–119
SAS Institute Inc (1988) SAS/STAT Users guide, release 6.03 ed. SAS Institute, Cary, North Carolina
SAS Institute Inc (1992) SAS technical report P-229. SAS/STAT software: changes and enhancements, release 6.07 ed. SAS Institute, Cary, North Carolina
Tinker NA, Mather DE (1995a) Methods for QTL analysis with progeny replicated in multiple environments. Jour Quant Trait Loci (on line). Avail. world wibe web: http://probe. nalusda.gov:8000/otherdocs/jqtl/jqtl1995-01/jqtl15.html
Tinker NA, Mather DE (1995b) MQTE: software for simplified composite interval mapping of QTLs in multiple environments. Jour Quant Trait Loci (on line). Avail. world wibe web: http//probe.nalusda.gov:8000/otherdocs/jqtl/jqtl 1995-02/jqtl16r2.html
Williams PT (1976) Pattern analysis in agricultural science. Elsevier Scientific Publishing Company, Amsterdam
Author information
Authors and Affiliations
Additional information
Communicated by J. W. Snape
Ignacio Romagosa was on sabbatical leave from the University of Lleida and the Institut de Recerca i Tecnologia Agroalimentàries, Lleida, Spain, when this study was conducted
Rights and permissions
About this article
Cite this article
Romagosa, I., Ullrich, S.E., Han, F. et al. Use of the additive main effects and multiplicative interaction model in QTL mapping for adaptation in barley. Theoret. Appl. Genetics 93, 30–37 (1996). https://doi.org/10.1007/BF00225723
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00225723