Abstract
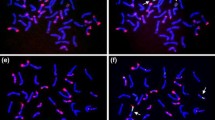
Copy numbers of four photosynthesis-related genes, PhyA, Ppc, RbcS and Lhcb1 *1, in wheat genomes were estimated by slot-blot analysis, and these genes were assigned to the chromosome arms of common wheat by Southern hybridization of DNA from an aneuploid series of the cultivar Chinese Spring. The copy number of PhyA was estimated to be one locus per haploid genome, and this gene was assigned to chromosomes 4AL, 4BS and 4DS. The Ppc gene showed a low copy number of small multigenes, and was located on the short arm of homoeologous group 3 chromosomes and the long arm of chromosomes of homoeologous group 7. RbcS consisted of a multigene family, with approximately 100 copies in the common wheat genome, and was located on the short arm of group 2 chromosomes and the long arm of group 5 chromosomes. Lhcb1 *1 also consisted of a multigene family with about 50 copies in common wheat. Only a limited number of restriction fragments (approximately 15%) were used to determine the locations of members of this family on the long arm of group 1 chromosomes owing to the multiplicity of DNA bands. The variability of hybridized bands with the four genes was less in polyploids, but was more in the case of multigene families. RFLP analysis of polyploid wheats and their presumed ancestors was carried out with probes of the oat PhyA gene, the maize Ppc gene, the wheat RbcS gene and the wheat Lhcb1 *1 gene. The RFLP patterns of common wheat most closely resembled those of T. Dicoccum (Emmer wheat), T. urartu (A genome), Ae. speltoides (S genome) and Ae. squarrosa (D genome). Diversification of genes in the wheat complex appear to have occurred mainly at the diploid level. Based on RFLP patterns, B and S genomes were clustered into two major groups. The fragment numbers per genome were reduced in proportion to the increase of ploidy level for all four genes, suggesting that some mechanism(s) might operate to restrict, and so keep to a minimum, the gene numbers in the polyploid genomes. However, the RbcS genes, located on 2BS, were more conserved (double dosage), indicating that the above mechanism(s) does not operate equally on individual genes.
Similar content being viewed by others
References
Ainsworth CC, Miller TE, Gale MD (1987) α-amylase and β-amylase homoeoloci in species related to wheat. Genet Res 49:93–103
Andreo CS, Gonzalez DH, Iglesias AA (1987) Higher plant phosphoenolpyruvate carboxylase. FEBS Eett 213:1–8
Appels R, Gerlach WR, Dennis ES, Swift H, Peacock WJ (1980) Molecular and chromosomal organization of DNA sequences coding for the ribosomal RNAs in cereals. Chromosoma 78:293–311
Bennet MD, Smith JB, Heslop-Harrison JS (1982) Nuclear amounts in angiosperms. Proc R Soc Lond B216:179–199
Brogue R, Coruzzi G, Lamppa G, Keith B, Chua NH (1983) Structural analysis of nuclear genes coding for the precursor to the small subunit of wheat ribulose-1,5-bisphosphate carboxylase. Bio/technology 1:55–61
Chang C, Bowman JL, DeJohn AW, Lander ES, Meyerowitz EM (1988) Restriction fragment length polymorphism linkage map for Arabidopsis thaliana. Proc Natl Acad Sci USA 85:6856–6860
Chao S, Raines CA, Longstaff M, Sharp PJ, Gale MD, Dyer TA (1989) Chromosomal location and copy number in wheat and some of its close relatives of genes for enzymes involved in photosynthesis. Mol Gen Genet 218:423–430
Chojecki AJS, Gale MD (1982) Genetic control of glucose phosphate isomerase in wheat and related species. Heredity 49:339–349
Christensen AH, Quail PH (1989) Structure and expression of a maize phytochrome-encoding gene. Gene 85:381–390
Cretin C, Santi S, Keryer E, Lepiniec L, Tagu D, Vidal J, Gadal P (1991) The phosphoenolpyruvate carboxylase gene family of Sorghum: promoter structures, amino-acid sequences and expression of genes. Gene 99:87–94
Dean C, Favreau M, Dunsmuir D, Bedbrook J (1987) Confirmation of the relative expression levels of the Petunia (Mitchell) rbc5 genes. Nucleic Acids Res 15:4655–4668
Dean C, Pichersky E, Dunsmuir D (1989) Structure, evolution, and regulation of RbcS genes in higher plants. Annu Rev Plant Physiol Plant Mol Biol 40:415–439
Dvorak J, McGuire PE, Cassidy B (1988) Apparent sources of the A genomes of wheats inferred from polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome 30:680–689
Ellis RJ (1979) The most abundant protein on earth. Trends Biochem Sci 4:241–244
Feinberg Vogelstein (1983) A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132:6–13
Fluhr R, Moses P, Morelli G, Coruzzi G, Chua NH (1986) Expression dynamics of the pea rbcS multigene family and organ distribution of the transcripts. EMBO J 5:2063–2071
Furuta Y, Nishikawa K, Yamaguchi S (1986) Nuclear DNA content in diploid wheat and its relatives in relation to the phylogeny of the tetraploid wheat. Jpn J Genet 61:97–105
Furuya M (1989) Molecular properties and biogenesis of phytochrome I and II. Adv Biophys 25:133–167
Galili S, Galili G, Feldman M (1991) Chromosomal location of genes for Rubisco small subunit and Rubisco-binding protein in common wheat. Theor Appl Genet 81:91–104
Galili S, Galili G, Avivi Y, Feldman M (1992) Identification and chromosomal location of four subfamilies of the rubisco small subunit genes in common wheat. Theor Appl Genet 83:385–391
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 9:1869–1886
Gilmartin PM, Sarokin L, Memelink J, Chua NH (1990) Molecular light switches for plant genes. Plant Cell 2:369–378
Green BR, Picherskey E, Kloppstech K (1991) Chlorophyll a/b-binding proteins: an extended gene family. Trends Biochem Sci 16:181–186
Heery DM, Gannon F, Powell R (1990) A simple method for subcloning DNA fragments from gel slices. Trends Genet 6:173
Hershey HP, Barker RF, Idler KB, Lissemore JL, Quail PH (1985) Analysis of cloned cDNA and genomic sequences for phyto-chrome: complete amino-acid sequences for two gene products expresses in etiolated Avena. Nucleic Acids Res 13:8543–8559
Izui K, Ishijima S, Yamaguchi Y, Katatiri F, Murata T, Shigesada K, Sugiyama T, Katsuki K (1986) Cloning and sequence analysis of cDNA encoding active phosphoenolpyruvate carboxylase of the C4-pathway from maize. Nucleic Acids Res 14:1615–1628
Jansson S, Pichersky E, Bassi R, Green BR, Ikeuchi M, Melis A, Simpson DJ, Spangfort M, Staehelin LA, Thornber JP (1992) A nomenclature for the genes encoding the chlorophyll a/b-binding proteins of higher plants. Plant Mol Biol Rep 10:242–253
Jellings AL, Leese BM, Leech RM (1983) Location of a chromosomal control of ribulose bisphosphate carboxylase amounts in wheat. Mol Gen Genet 192:272–274
Kawamura T, Shigesada K, Yanagisawa S, Izui K (1990) Phosphoenolpyruvate carboxylase prevalent in maize roots: isolation of a cDNA clone and its use for analyses of the gene and gene expression. J Biochem 107:165–168
Kay S, Keith B, Shinozaki K, Chye ML, Chua NH (1989) The rice phytochrome gene: structure, autoregulated expression and binding of GT-1 to a conserved site in the 5′ upstream region. The Plant Cell 1:353–360
Kihara H (1944) Die Entdeckung der DD-Analysatoren beim Weizen. Agr Hort Tokyo 19:889–890
Kihara H (1954) Consideration on the evolution and distribution of Aegilops species based on the analyzer-method. Cytologia 19:336–357
Lamppa GK, Morelli G, Chua NH (1985) Structure and developmental regulation of a wheat gene encoding the major chlorophyll a/b-binding polypeptide. Mol Cell Biol 5:1370–1378
Latzko E, Kelly GJ (1983) The many-faceted function of phosphoenolpyruvate carboxylase in C3 plants. Physiol Veg 21:805–808
Luan S, Bogorad L (1989) Nucleotide sequences of the two genes encoding the light harvesting chlorophyll a/b-binding protein of rice. Nucleic Acids Res 17:2357–2358
Maniatis T, Sambrook J, Fritsch EF (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Martin G, O'Dell M, Flavell RB (1982) Partial innactivation of wheat nucleolus organizers by the nucleolus organizer chromosomes from Aegilops umbellulata. Chromosoma 84:687–700
Matsuoka M, Yamamoto N (1989) Induction of mRNAs for phosphoenolpyruvate carboxylase and pyruvate, orthophosphate dikinase in leaves of a C3 plant exposed to light. Plant Cell Physiol 30:479–486
McFadden ES, Sears ER (1944) The artificial synthesis of Triticum spelta. Rec Genet Soc America 13:26–27
McIntosh L, Poulsen C, Bogorad L (1980) Chloroplast gene sequence for the large subunit of ribulose bisphosphate carboxylases of maize. Nature 288:556–560
Miziorko HM, Lorimer GH (1983) Ribulose-1, 5-bisphosphate carboxylase-oxygenase. Annu Rev Biochem 52:507–535
Murray HG, Thompson WF (1980) Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res 8:4321–4325
Nishikawa K (1983) Species relationship of wheat and its putative ancestors as viewed from isozyme variations. Proc 6th Int Wheat Genet Symp, Kyoto, pp 59–63
Ogihara Y, Tsunewaki K (1988) Diversity and evolution of chloroplast DNA in Triticum and Aegilops. Theor Appl Genet 76:321–332
O'Leary MH (1982) Phosphoenolpyruvate carboxylase: an enzymologist's view. Annu Rev Plant Physiol 33:297–315
Pichersky E, Subramaniam R, White MJ, Reid J, Aebersold R, Green BR (1991) Chlorophyll a/b-binding (CAB) polypeptides of CP29, the internal chlorophyll a/b complex of PSII: Characterization of the tomato gene encoding the 26-kDa (type I) polypeptide, and evidence for a second CP29 polypeptide. Mol Gen Genet 227:277–284
Quail PH (1991) Phytochrome: a light-activated molecular switch that regulates plant gene expression. Annu Rev Genet 25:389–409
Quail PH, Hershey HP, Idler KB, Sarrock RA, Christensen AH (1991) Phytochrome properties and biological action. Springer Verlag, Berlin
Sato N (1988) Nucleotide sequence and expression of the phytochrome gene in Pisum sativum: differential regulation by light of multiple transcript. Plant Mol Biol 11:697–710
Sharrock RA, Quail PH (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3:1745–1757
Sheen JY, Bogorad L (1987) Differential expression of C4 pathway genes in mesophyll and bundle sheath cells of greening maize leaves. J Biol Chem 262:11726–11730
Stayton MM, Black M, Bedbrook J, Dunsmuir P (1986) A novel chlorophyll a/b (Cab)-protein gene from petunia which encodes the lower molecular weight Cab precursor protein. Nucleic Acids Res 14:9781–9796
Sugita M, Gruissem W (1987) Developmental, organ-specific, and light-dependent expression of the tomato ribulose-1,5-bisphosphate carboxylase small subunit gene family. Proc Natl Acad Sci USA 84:7104–7108
Terachi T, Tsunewaki K (1992) The molecular basis of genetic diversity among cytoplasms of Triticum and Aegilops. VII. Mitochondrial RFLP analysis using cloned genes as probes. Mol Biol Evol 9:917–931
Thompson RD, Bartels D, Harberd HP, Flavell RB (1983) Characterization of the multigene family coding for HMW gultenin subunits in wheat using cDNA clones. Theor Appl Genet 67:87–96
Ting IP, Osmond CB (1973) Multiforms of plant phosphoenolpyruvate carboxylase associated with different metabolic pathways. Plant Physiol 51:448–453
Tsunewaki K, Takumi S, Mori N, Achiwa T, Liu YG (1992) Origin of polyploid wheats revealed by RFLP analyses, In: Sasakuma T, Kinoshita T (eds) Proc Dr H Kihara Mem Int Sym on Cytoplasmic Engineering in Wheat. Kihara Mem Found, Yokohama, pp 31–39
Walling LL, Chang YC, Demmin DS, Holzer FM (1988) Isolation, characterization and evolutionary relatedness of three members from the soybean mutigene family encoding chlorophyll a/b-binding proteins. Nucleic Acids Res 16:10477–10492
Wolter FP, Fritz CC, Willmitzer L, Schell J, Schreier PH (1988) The rbcS gene in Solanum tuberosum: conservation of transit peptide and exon shuffling during evolution. Proc Natl Acad Sci USA 85:846–850
Author information
Authors and Affiliations
Additional information
Communicated by J. Mac Key
Rights and permissions
About this article
Cite this article
Ogihara, Y., Shimizu, H., Hasegawa, K. et al. Chromosome assignment of four photosynthesis-related genes and their variability in wheat species. Theoret. Appl. Genetics 88, 383–394 (1994). https://doi.org/10.1007/BF00223649
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00223649