Abstract
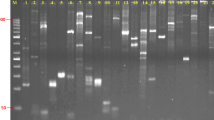
Several techniques of DNA analysis were applied to identify chrysanthemum cultivars. Unrelated cultivars could be distinguished by using RAPDs (random amplified polymorphic DNAs), inter-SSR (simple sequence repeat) PCR (polymerase chain reaction), hybridization-based DNA fingerprinting, as well as RFLPs (restriction fragment length polymorphisms). Cultivars with different flower colours and belonging to one family, i.e. vegetatively derived from 1 cultivar, appeared to have the same DNA fragment patterns, whichever technique was applied. The absence of polymorphisms between different accessions of the same cultivar indicated a high stability of the observed patterns.
Similar content being viewed by others
References
Anderson NO, Ascher PD, Widmer RE (1992) Inbreeding depression in garden and glasshouse chrysanthemums: germination and survivorship. Euphytica 62:155–169
Baird E, Cooperbland S, Waugh R, Demaine M, Powell W (1992) Molecular characterisation of inter-specific and intra-specific somatic hybrids of potato using randomly amplified polymorphic DNA (RAPD) markers. Mol Gen Genet 233:469–475
Broertjes C, Van Harten AM (1978) Application of mutation breeding methods in the improvement of vegetatively propagated crops. Elsevier, Amsterdam
Brown PTH, Lörz H (1986) Molecular changes and possible origins of somaclonal variation. In: Semal J (ed) Somaclonal variations and crop improvement. Martinus Nijhoff, Boston, pp. 148–159
Brown PTH, Lange FD, Kranz E, Lörz H (1993) Analysis of single protoplasts and regenerated plants by PCR and RAPD technology. Mol Gen Genet 237:311–317
Bush SR, Earle ED, Langhans RW (1976) Plantlets from petal segments, petal epidermis, and shoot tips of the periclinal chimera, Chrysanthemum morifolium ‘Indianapolis’ Am J Bot 63:729–737
Caetano-Anollés G, Bassam BJ, Gresshoff PM (1991) DNA amplification fingerprinting using very short arbitrary oligonucleotide primers. Bio/Technology 9:553–557
Demeke T, Kawchuk LM, Lynch DR (1993) Identification of potato cultivars and clonal variants by random amplified polymorphic DNA analysis. Am Potato J 70:561–570
Dowrick GJ, El-Bayoumi A (1966) The origin of new forms of the garden Chrysanthemum. Euphytica 15:32–38
Fiebich D, Hennig F (1992) Use of isozyme analysis in breeding of chrysanthemum. Gartenbauwissenschaft 57:212–218
Gorg R, Schachtschabel U, Ritter E, Salamini F, Gebhardt C (1992) Discrimination among 136 tetraploid potato varieties by fingerprints using highly polymorphic DNA markers. Crop Sci 32:815–819
Graham J, McNicol RJ, Greig K, Van De Ven WTG (1994) Identification of red raspberry cultivars and an assessment of their relatedness using fingerprints produced by random primers. J Hortic Sci 69:123–130
Hu J, Quiros CF (1991) Identification of broccoli and cauliflower cultivars with RAPD markers. Plant Cell Rep 10:505–511
Isabel N, Tremblay L, Michaud M, Tremblay FM, Bousquet J (1993) RAPDs as an aid to evaluate the genetic integrity of somatic embryogenesis-derived populations of Picea mariana (Mill.) B.S.P. Theor Appl Genet 86:81–87
Koller B, Lehmann A, McDermott JM, Gessler C (1993) Identification of apple cultivars using RAPD markers. Theor Appl Genet 85:901–904
Lavi U, Hillel J, Lahav E, Sharon D (1991) Application of DNA fingerprints for identification and genetic analysis of avacado. J Am Soc Hortic Sci 116:1078–1081
Lee M, Phillips RL (1988) The chromosomal basis of somaclonal variation. Annu Rev Plant Physiol 39:413–437
Mailer RJ, Scarth R, Fristensky B (1994) Discrimination among cultivars of rapeseed (Brassica napus L.) using DNA polymorphisms amplified from arbitrary primers. Theor Appl Genet 87:697–704
Malaure RS, Barclay G, Power JB, Davey MR (1991a) The production of novel plants from florets of Chrysanthemum morifolium using tissue culture. 1. Shoot regeneration from ray florets and somaclonal variation exhibited by the regenerated plants. J Plant Physiol 139:8–13
Malaure RS, Barclay G, Power JB, Davey MR (1991b) The production of novel plants from florets of Chrysanthemum morifolium using tissue culture. 2. Securing natural mutations (sports.) J Plant Physiol 139:14–18
Marsolais JV, Pringle JS, White BN (1993) Assessment of random amplified polymorphic DNA (RAPD) as genetic markers for determining the origin of interspecific lilac hybrids. Taxon 42:531–537
Mulcahy DL, Cresti M, Sansavini S, Douglas GC, Linskens HF, Mulcahy GB, Vignani R, Pancaldi M (1993) The use of Random Amplified Polymorphic DNAs to fingerprint apple genotypes. Sci Hortic 54:89–96
Nilke M, Nowak J, Wright JM, Mclean NL (1993) DNA fingerprinting of red clover (Trifolium pratense L) with Jeffrey probes —detection of somaclonal variation and other applications. Plant Cell Rep 13:72–78
Nybom H, Schaal BA, Rogstad SH (1989) DNA “fingerprints” can distinguish cultivars of blackberries and rapsberries. Acta Hortic 262:305–310
Parent JG, Fortin MG, Page D (1993) Identification of raspberry cultivars by random amplified polymorphic DNA (RAPD) analysis. Can J Plant Sci 73:1115–1122
Poulsen GB, Kahl G, Weising K (1993) Oligonucleotide fingerprinting of resynthesized Brassica napus. Euphytica 70:53–59
Rajapakse S, Hubbard M, Kelly JW, Abbott AG, Ballard RE (1992) Identification of rose cultivars by restriction fragment length polymorphism. Sci Hortic 52:237–245
Stewart RN, Dermen H (1970) Somatic genetic analysis of the apical layers of chimeral sports in Chrysanthemum by experimental production of adventitious shoots. Am J Bot 57:1061–1971
Striem MJ, Spiegel-Roy P, Ben-Hayyim G, Beckmann J, Gidoni D (1990) Genomic DNA fingerprinting of Vitis vinifera by the use of multi-loci probes. Vitis 29:223–227
Teynor TM, Ascher PD, Widmer RE, Luby JJ (1989) Inheritance of flower color in Dendranthema grandiflora Tzvelev. (Chrysanthemum morifolium Ramat.) using cultivars and inbreds. II Vacuole pigmentation. Euphytica 42:297–305
Thomas MR, Matsumoto S, Cain P, Scott NS (1993) Repetitive DNA of grapevine:classes present and sequences suitable for cultivar identification. Theor Appl Genet 86:173–180
Vallés MP, Wang ZY, Montavon P, Potrykus I, Spangenberg G (1993) Analysis of genetic stability of plants regenerated from suspension cultures and protoplasts of meadow fescue (Festuca pratensis Huds.) Plant Cell Rep 12:101–106
Vosman B, Arens P, Rus-Kortekaas W, Smulders MJM (1992) Identification of highly polymorphic DNA regions in tomato. Theor Appl Genet 85:239–244
Watanabe K (1977) The control of diploidlike meiosis in polyploid taxa of Chrysanthemum (Compositae). Jpn J Genet 52: 125–131
Weising K, Nybom H, Meyer W, Wolff K (1994) DNA fingerprinting in plants and fungi. CRC Press, Florida
Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Rec 18:7213–7218
Wilde J, Waugh R, Powell W (1992) Genetic fingerprinting of Theobroma clones using randomly amplified polymorphic DNA markers. Theor Appl Genet 83:871–877
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Wolff K, Peters-Van Rijn J (1993) Rapid detection of genetic variability in chrysanthemum (Dendranthema grandiflora Tzvelev) using random primers. Heredity 71:335–341
Wolff K, Rogstad SH, Schaal BA (1994a) Population and species variation of minisatellite DNA in Plantago. Theor Appl Genet 87:733–740
Wolff K, Peters-Van Rijn J, Hofstra H (1994b) RFLP analysis in chrysanthemum. I. Probe and primer development. Theor Appl Genet 88:472–478
Wu L, Lin H (1994) Identifying buffalograss [Buchloe dactyloides (Nutt) Engelm] cultivar breeding lines using random amplified polymorphic DNA (RAPD) markers. J Am Soc Hortic Sci 119:126–130
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR) —anchored polymerase chain reaction amplification. Genomics 20:176–183
Author information
Authors and Affiliations
Additional information
Communicated by P. M. A. Tigerstedt
Rights and permissions
About this article
Cite this article
Wolff, K., Zietkiewicz, E. & Hofstra, H. Identification of chrysanthemum cultivars and stability of DNA fingerprint patterns. Theoret. Appl. Genetics 91, 439–447 (1995). https://doi.org/10.1007/BF00222971
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00222971